ONLINE INQUIRY
- Specification
- Background
- Scientific Data
- Q & A
- Customer Review
Conduct studies of metabolic stability (% of original drug remaining over time)
Identify which cytochrome P450 (CYP) enzymes oxidize a drug candidate and which UDP-glucuronosyltransferase (UGT) enzymes are responsible for glucuronidation
Identify, quantify and characterize drug metabolites
Describe a drug's metabolic pathways
Identify inhibition of cytochrome P450 (CYP) enzymes
Human liver microsomes (HLMs) are approximately spherical membrane vesicles formed by the self-fusion of fragmented endoplasmic reticulum. They are isolated from liver tissue and separated by differential centrifugation. These structures contain membrane proteins, phospholipids, and other proteins. "50-donors" means a concentrated preparation of liver tissue from 50 donors. This microsomes-50 donor provides a median metabolic profile from a population, thus removing individual differences in the study of drug metabolism.
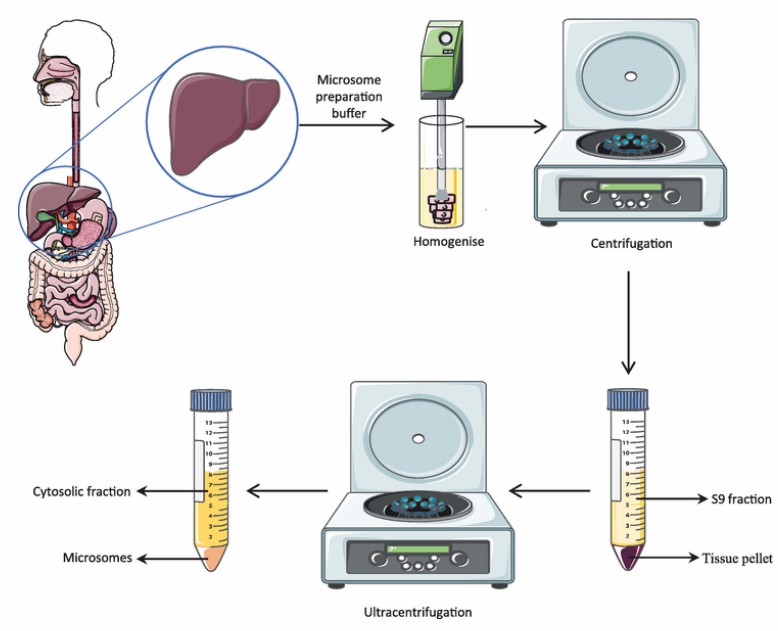 Fig. 1. Preparation of human liver microsomes (Rao Gajula, S. N., Vora S. A., et al., 2021).
Fig. 1. Preparation of human liver microsomes (Rao Gajula, S. N., Vora S. A., et al., 2021).
The key functional units in HLMs are the enzyme proteins. They include Phase I metabolic enzymes, like the cytochrome P450 family, and Phase II metabolic enzymes, including uridine diphosphate glucuronosyltransferases (UGTs) and sulfotransferases (STs), flavin-containing monooxygenases (FMOs), fatty acid-CoA synthetases, alcohol dehydrogenases, carboxylesterases, etc. These enzymes are responsible for reactions such as hydrolysis, oxidation, reduction, and conjugation, which can break down toxic substances or convert non-toxic chemicals into toxic or carcinogenic intermediates.
HLMs are widely employed for high-throughput drug metabolism stability studies, evaluating the metabolic rate and pathways of drug candidates, drug-drug interactions, and individual drug clearance. Among the CYP450 enzymes studied, NADPH or an NADPH-regenerating system can trigger the reaction. For UGT enzyme activity studies, uridine diphosphate glucuronic acid (UDPGA) combined with alamethicin initiates the reaction. So, in vitro drug metabolism using human liver microsomes can be convenient, easy and highly stable. In addition, they can keep enzyme activity stable for years in frozen forms, so they are easy to store. Thus, liver microsomes have become an essential tool in in vitro drug metabolism studies.
The Metabolism of Gelsevirine in Human, Pig, Goat and Rat Liver Microsomes
Gelsemium is a small genus of flowering plants from the family Loganiaceae comprising five species, three of which, Gelsemium sempervirens (L.) J. St.-Hil., G. elegans Benth and G. rankinii Small, are particularly popular. Although the pharmacological activities and metabolism of koumine and gelsemine have been reported in previous studies, the species differences of gelsevirine metabolism have not been well studied. The aim of the present work was to investigate the metabolism of gelsevirine in liver microsomes of humans, pigs, goats and rats using HPLC-QqTOF/MS.
Accurate extracted ion chromatograms (EICs) of unchanged gelsevirine and its metabolites formed in human liver microsomes in the presence of the NADPH-generating system are shown in Figure 1. In addition to the parent drug, a total of six metabolites were detected in human liver microsomes. By analyzing the ion fragments, the species of M1-6 were predicted. M1 was tentatively identified as N-demethyl-gelsevirine. M2 was a hydrogenated metabolite of gelsevirine at the C = C double bond. M3 was tentatively identified as the gelsevirine N-oxide on the nitrogen at position 4. M4 was a hydroxylated and hydrogenated metabolite of gelsevirine. M5 was a dehydrogenated metabolite of gelsevirine at carbon 21 and nitrogen 4. M6 is a hydroxylated metabolite of M5 (Fig. 1).
The analysis of gelatinvirine metabolites in liver microsomes of humans, pigs, goats, and rats revealed consistent detection of metabolites M1 to M4 in both rat and goat liver microsomes. Additionally, human liver microsomes also contained metabolites M5 and M6. Pig liver microsomes revealed five metabolites (M1, M3-M6). The intensity of metabolite M1 was about ten times higher in pigs than in the other species. Metabolite M3 levels were double in pig and goat liver microsomes compared to human and rat liver microsomes. Metabolite M1 was present in all species in the order: pig > human > goat > rat. Metabolite M2 was not found in pig liver microsomes but was most abundant in humans, followed by rats and goats. Metabolite M3 was highest in pig liver microsomes and lowest in rat liver microsomes. Similar levels of M4 were found in pig, goat, and rat liver microsomes, with slightly higher amounts in humans. Metabolites M5 and M6 were unique to pig and human liver microsomes, being more abundant in pigs. Based on these findings, the metabolic pathways of gelatinvirine in liver microsomes were proposed and are illustrated in Figure 2.
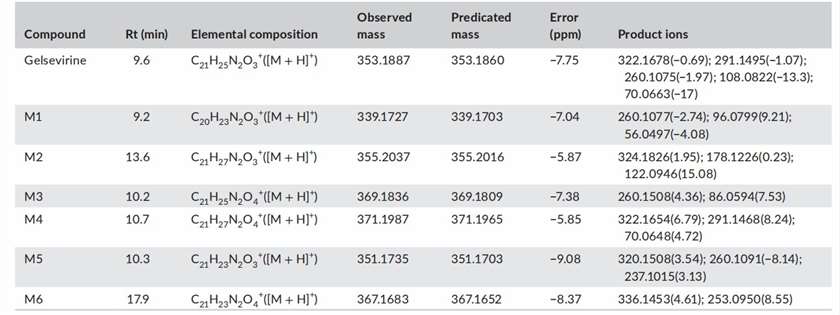 Fig. 1. Retention times (Rts), elemental compositions, observed and predicted masses, mass errors and product ions of gelsevirine and its metabolites (Zhang, H. H., Yang, W. J., et al., 2021).
Fig. 1. Retention times (Rts), elemental compositions, observed and predicted masses, mass errors and product ions of gelsevirine and its metabolites (Zhang, H. H., Yang, W. J., et al., 2021).
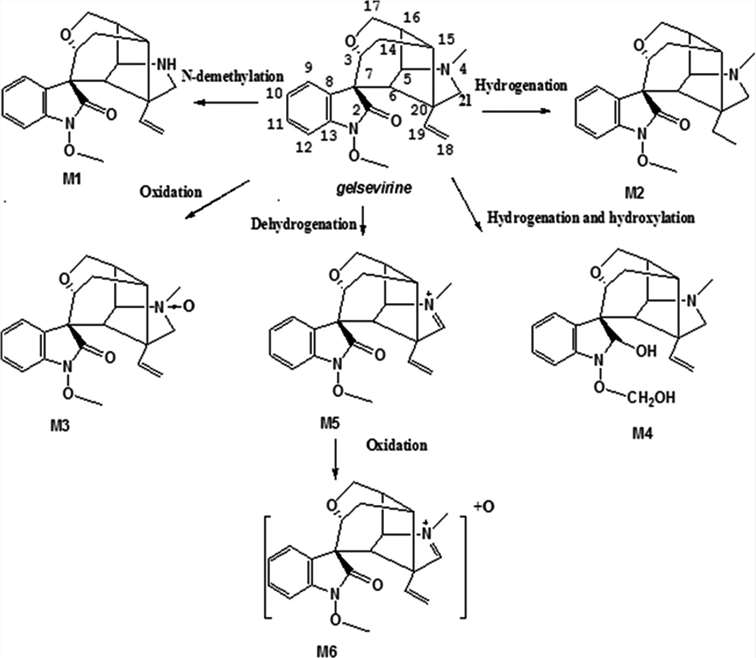 Fig. 2. Proposed metabolic pathways of gelsevirine in liver microsomes in vitro (Zhang, H. H., Yang, W. J., et al., 2021).
Fig. 2. Proposed metabolic pathways of gelsevirine in liver microsomes in vitro (Zhang, H. H., Yang, W. J., et al., 2021).
Biotransformation of the New Synthetic Cannabinoid with an Alkene, MDMB-4en-PINACA, by Human Hepatocytes, Human Liver Microsomes, and Human Urine and Blood
This study reports in vitro and in vivo metabolism of a new synthetic cannabinoid, methyl 3,3-dimethyl-2-[1-(pent-4-en-1-yl)-1H-indazole-3-carboxamido] butanoate (MDMB-4en-PINACA). MDMB-4en-PINACA was incubated with both human hepatocytes and human liver microsomes (HLM) for up to 5 h and 1 h, respectively. An authentic human urine and a corresponding blood sample were analyzed to confirm the in vitro metabolites.
In vitro studies resulted in the detection of 11 metabolites for hepatocyte incubations and 31 metabolites for HLM incubations, of which 10 metabolites were common metabolites with hepatocytes (Fig. 3). The metabolites were presumably formed by butanoic acid formation, carboxylation, dehydrogenation, dihydrodiol formation, ester hydrolysis, hydrogenation, hydroxylation, and glucuronidation either alone or in combination. The only phase II metaboliteobserved was M4 (glucuronide of M8, ester hydrolysis, and dihydrodiol). The metabolites eluted between 3.60 and 10.11 min before the parent drug eluting at 11.66 min (Fig. 4). The three most abundant metabolites were M8 (ester hydrolysis and dihydrodiol), M30 (ester hydrolysis), and M20 (ester hydrolysis and hydroxylation) for 5 h hepatocyte incubation and M7 (ester hydrolysis, dihydrodiol, and dehydrogenation), M8 (ester hydrolysis and dihydrodiol), and M3 (ester hydrolysis, dihydrodiol, hydroxylation, and dehydrogenation) for HLM incubation.
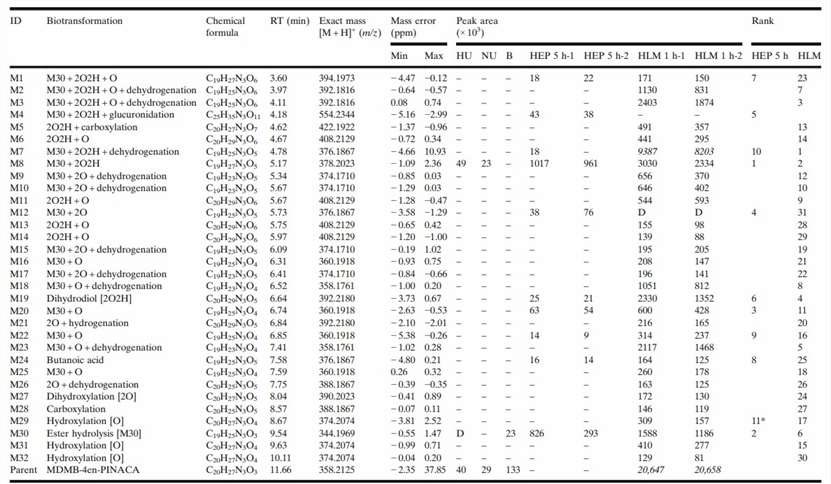 Fig. 3. MDMB-4en-PINACA Metabolites with Biotransformations, Chemical Formulas, Retention Times, Exact Masses of the Protonated Molecules, Mass Errors (Watanabe, S., Vikingsson, S., et al., 2019).
Fig. 3. MDMB-4en-PINACA Metabolites with Biotransformations, Chemical Formulas, Retention Times, Exact Masses of the Protonated Molecules, Mass Errors (Watanabe, S., Vikingsson, S., et al., 2019).
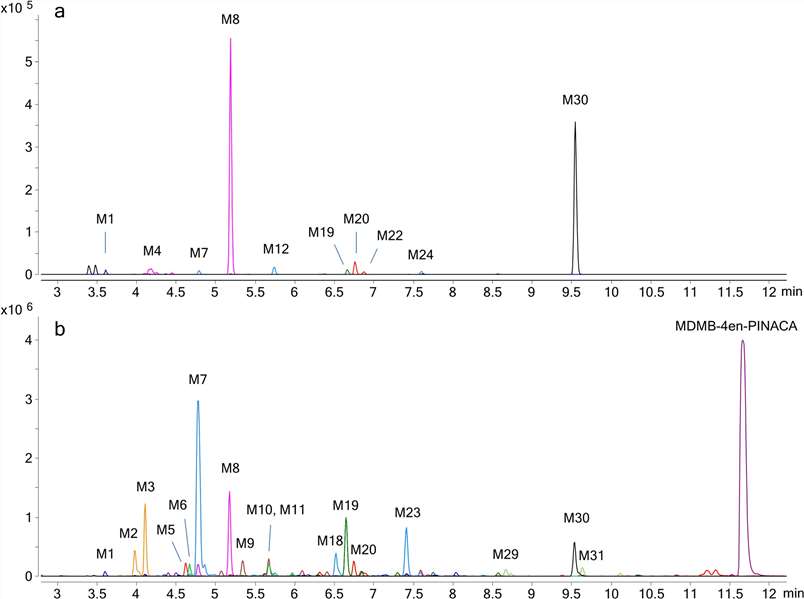 Fig. 4. Combined extracted ion chromatograms of MDMB-4en-PINACA metabolites found in a human hepatocytes and b HLM with labels for larger peaks (Watanabe, S., Vikingsson, S., et al., 2019).
Fig. 4. Combined extracted ion chromatograms of MDMB-4en-PINACA metabolites found in a human hepatocytes and b HLM with labels for larger peaks (Watanabe, S., Vikingsson, S., et al., 2019).
Ask a Question
Write your own review