FISH-Based Clonality Analysis Services
- Service Details
- Features
- Case Studies
- FAQ
- Explore Other Options
A well-characterized clonally derived cell line is not only a fundamental component of the biopharmaceutical manufacturing process, but also a requirement for regulatory submission, most commonly at the Investigational New Drug (IND) stage. Clonality is thought to minimize the heterogeneity of cell banks and thus allow for the consistent manufacture of a product. Regulatory agencies require that all biomanufacturing cell lines be "cloned from a single cell progenitor" as part of the overall control strategy to ensure safety and consistent product quality. The United States (FDA) and the European Union (EMA) have different, but overlapping, guidelines for the manufacture of biotherapeutics regarding the clonal derivation of production cell lines.
Recent developments such as single-cell sorting and high-throughput imaging allow for the acquisition of evidence supporting clonality. Rederiving a clonal cell line by additional limited dilutions is time-consuming, costly, and, crucially, may affect the production and growth rates of cell lines. This is especially undesired when timelines for submission are short. Creative Bioarray offers comprehensive fluorescence in situ hybridization (FISH)-based clonality analysis services to support our clients in maintaining the highest standards of product quality and regulatory compliance.
Why Choose FISH for Clonality Analysis?
- FISH techniques have a high degree of sensitivity, allowing the detection of monoclonal cells within a sample.
- FISH allows for the visual identification and quantification of monoclonal cells within a sample.
- FISH can be performed on a wide range of sample types.
Comprehensive Solutions for FISH-Based Clonality Analysis
FISH-Based Clonality Analysis for MCB, WCB, and EOPC
The success of biologics manufacturing hinges on the genetic integrity of the production cell line. Creative Bioarray's FISH-based clonality analysis services are designed to thoroughly characterize the genetic profile of your Master Cell Bank (MCB), Working Cell Bank (WCB), and End-of-Production Cells (EOPC).
Our expert team utilizes advanced FISH techniques to assess the chromosome number, identify any chromosomal abnormalities or rearrangements, and determine the clonality of cell lines. We also provide detailed reports on the genetic composition and stability of cell lines throughout the manufacturing process, ensuring the consistent quality and safety of your biotherapeutic products.
FISH-Based Clonality Analysis for different cell lines (CHO, HEK293)
Our FISH-based clonality analysis services are designed to accommodate a wide range of cell lines, including the industry-leading Chinese Hamster Ovary (CHO) and Human Embryonic Kidney (HEK293) cell lines. Whether you are working with a newly developed cell line or an established one, our team of specialists will work closely with you to optimize the FISH analysis protocols and provide comprehensive insights into the genetic profile and clonality of your cells. Our comprehensive services involve chromosome preparation, FISH probe design and synthesis, and FISH analysis.
Creative Bioarray's Clonality Analysis Service (FISH) Has the Following Features
- Detection of each integration site
- Analyze 100-200 cells per sample
- Rapid 4-week turnaround time, including additional reporting
- High accuracy and sensitively
- Competitive pricing
Case Studies
Case 1: FISH-Based Clonality Analysis for MCB
Chromosomal rearrangements (or genetic plasticity) would occur in CHO cells over a 30-day period of culture, which is the case with an MCB. Out of the 200 cells that were randomly captured and analyzed, 200 cells (100%) showed uniform single-site integration in the centromeric region of Chr.10.
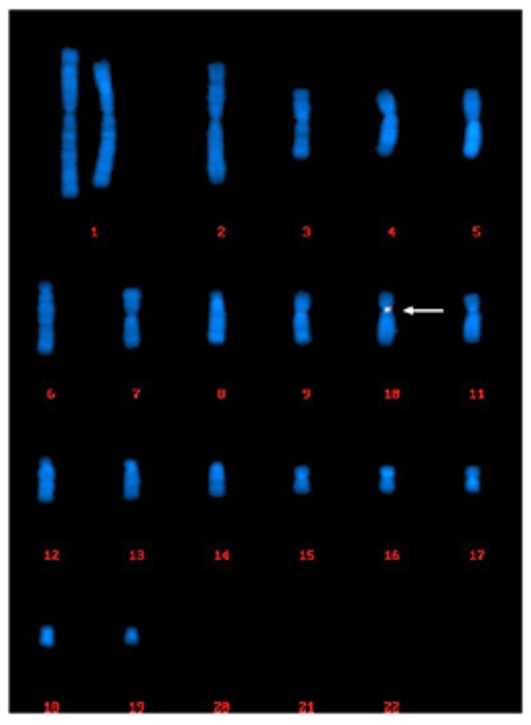 Fig. 1 (200/200 cells) Centromeric region of chr.10.
Fig. 1 (200/200 cells) Centromeric region of chr.10.
Case 2: FISH-Based Clonality Analysis for F7C2-P3C63 Cell Line
In 95% of the cell population analyzed, we have identified a dominant pattern characterized by the presence of FISH signals on two chromosomes, which is the long arm of the large submetacentric chromosome, positioned at #1 on the karyograms and the long arm of the medium-sized submetacentric chromosome, positioned at #6 on the karyograms (95 out of 100 cells). Apart from the dominant pattern, another two patterns were observed. In four cells, FISH signals were observed only on chromosome 1. In one cell, FISH signals were observed on chromosome 6 and an unknown chromosome.
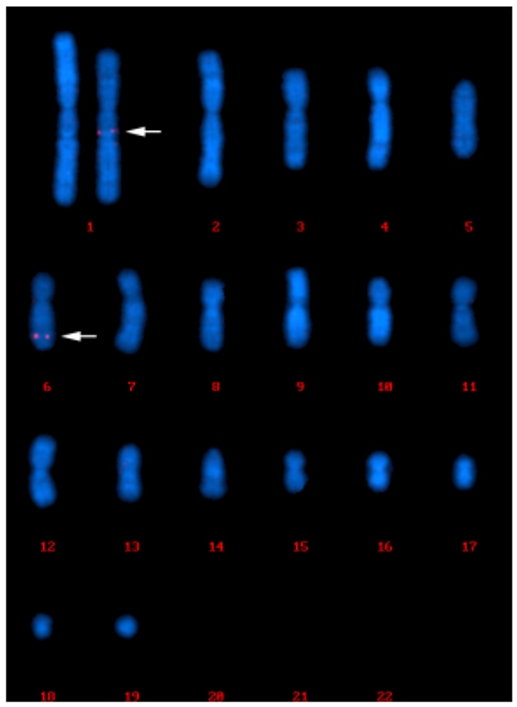 Fig. 2 FISH signals were observed on chromosome 1 and chromosome 6.
Fig. 2 FISH signals were observed on chromosome 1 and chromosome 6.
FAQ
1. How does FISH-based clonality analysis work?
FISH-based clonality analysis works by hybridizing fluorescently labeled DNA probes to specific target sequences within the cells' chromosomes. By visualizing these probes under a fluorescence microscope, researchers can identify and quantify the presence of specific genetic markers. Consistent patterns across multiple cells indicate clonality, whereas heterogeneous patterns suggest a polyclonal or mixed-cell population.
2. What are the advantages of using FISH for clonality analysis compared to other methods?
The advantages of using FISH for clonality analysis include high specificity and sensitivity for detecting genetic markers, the ability to visualize genetic changes at the single-cell level, relatively rapid and straightforward procedure compared to molecular techniques like PCR or next-generation sequencing, the capability to detect both large chromosomal abnormalities and smaller genetic changes.
3. What are the applications of FISH-based clonality analysis?
Confirming the monoclonality of cell lines used in research and biopharmaceutical production; assessing the clonal evolution of cancer cells during treatment; and ensuring the genetic uniformity of genetically engineered cell lines for therapeutic purposes.
At Creative Bioarray, our FISH-based clonality analysis services are designed to meet the stringent requirements of regulatory bodies, such as the FDA and EMA, ensuring that your data is aligned with the necessary guidelines. If you have any special needs or questions regarding our services, please feel free to contact us or make an online inquiry.
References
- Frye, Christopher, et al. "Industry view on the relative importance of "clonality" of biopharmaceutical-producing cell lines." Biologicals 44.2 (2016): 117-122.
- Wurm, Florian M., and Maria João Wurm. "Cloning of CHO cells, productivity, and genetic stability-a discussion." Processes 5.2 (2017): 20.
- ICH Q5D Derivation and characterization of cell substrates used for the production of biotechnological/biological products (CPMP/ICH/294/95).
- Welch, J. (2017). Tilting at clones: A regulatory perspective on the importance of "Clonality" of mammalian cell banks. CDER/OPQ/OBP/DBRRIV April 24, 2017.
Explore Other Options

