- You are here: Home
- Services
- ISH/FISH Services
- FISH Applications
- Splice Variant Analysis (FISH)
Services
-
Cell Services
- Cell Line Authentication
- Cell Surface Marker Validation Service
-
Cell Line Testing and Assays
- Toxicology Assay
- Drug-Resistant Cell Models
- Cell Viability Assays
- Cell Proliferation Assays
- Cell Migration Assays
- Soft Agar Colony Formation Assay Service
- SRB Assay
- Cell Apoptosis Assays
- Cell Cycle Assays
- Cell Angiogenesis Assays
- DNA/RNA Extraction
- Custom Cell & Tissue Lysate Service
- Cellular Phosphorylation Assays
- Stability Testing
- Sterility Testing
- Endotoxin Detection and Removal
- Phagocytosis Assays
- Cell-Based Screening and Profiling Services
- 3D-Based Services
- Custom Cell Services
- Cell-based LNP Evaluation
-
Stem Cell Research
- iPSC Generation
- iPSC Characterization
-
iPSC Differentiation
- Neural Stem Cells Differentiation Service from iPSC
- Astrocyte Differentiation Service from iPSC
- Retinal Pigment Epithelium (RPE) Differentiation Service from iPSC
- Cardiomyocyte Differentiation Service from iPSC
- T Cell, NK Cell Differentiation Service from iPSC
- Hepatocyte Differentiation Service from iPSC
- Beta Cell Differentiation Service from iPSC
- Brain Organoid Differentiation Service from iPSC
- Cardiac Organoid Differentiation Service from iPSC
- Kidney Organoid Differentiation Service from iPSC
- GABAnergic Neuron Differentiation Service from iPSC
- Undifferentiated iPSC Detection
- iPSC Gene Editing
- iPSC Expanding Service
- MSC Services
- Stem Cell Assay Development and Screening
- Cell Immortalization
-
ISH/FISH Services
- In Situ Hybridization (ISH) & RNAscope Service
- Fluorescent In Situ Hybridization
- FISH Probe Design, Synthesis and Testing Service
-
FISH Applications
- Multicolor FISH (M-FISH) Analysis
- Chromosome Analysis of ES and iPS Cells
- RNA FISH in Plant Service
- Mouse Model and PDX Analysis (FISH)
- Cell Transplantation Analysis (FISH)
- In Situ Detection of CAR-T Cells & Oncolytic Viruses
- CAR-T/CAR-NK Target Assessment Service (ISH)
- ImmunoFISH Analysis (FISH+IHC)
- Splice Variant Analysis (FISH)
- Telomere Length Analysis (Q-FISH)
- Telomere Length Analysis (qPCR assay)
- FISH Analysis of Microorganisms
- Neoplasms FISH Analysis
- CARD-FISH for Environmental Microorganisms (FISH)
- FISH Quality Control Services
- QuantiGene Plex Assay
- Circulating Tumor Cell (CTC) FISH
- mtRNA Analysis (FISH)
- In Situ Detection of Chemokines/Cytokines
- In Situ Detection of Virus
- Transgene Mapping (FISH)
- Transgene Mapping (Locus Amplification & Sequencing)
- Stable Cell Line Genetic Stability Testing
- Genetic Stability Testing (Locus Amplification & Sequencing + ddPCR)
- Clonality Analysis Service (FISH)
- Karyotyping (G-banded) Service
- Animal Chromosome Analysis (G-banded) Service
- I-FISH Service
- AAV Biodistribution Analysis (RNA ISH)
- Molecular Karyotyping (aCGH)
- Droplet Digital PCR (ddPCR) Service
- Digital ISH Image Quantification and Statistical Analysis
- SCE (Sister Chromatid Exchange) Analysis
- Biosample Services
- Histology Services
- Exosome Research Services
- In Vitro DMPK Services
-
In Vivo DMPK Services
- Pharmacokinetic and Toxicokinetic
- PK/PD Biomarker Analysis
- Bioavailability and Bioequivalence
- Bioanalytical Package
- Metabolite Profiling and Identification
- In Vivo Toxicity Study
- Mass Balance, Excretion and Expired Air Collection
- Administration Routes and Biofluid Sampling
- Quantitative Tissue Distribution
- Target Tissue Exposure
- In Vivo Blood-Brain-Barrier Assay
- Drug Toxicity Services
Splice Variant Analysis (FISH)
Alternative splicing is a fundamental regulatory process of gene expression that allows generation of multiple mRNA isoforms from a single gene, which plays significant roles in human disease, particularly cancer and neurological disorders. Splice variants that are found predominantly in tumors have clear diagnostic value and may provide potential drug targets. And understanding the process of aberrant splicing and the characterization of the splice variants may prove curcial to our understanding of malignant transformation.
Currently, there is only a limited number of methods have been explored to detect RNA splicing variants. Detection of short isoform-specific sequences requires RNA isolation for PCR analysis – an approach that loses the reginal and cell-type-specific distribution of isoforms. Having the capability to distinguish the differential expression of RNA variants in tissue is critical because alterations in mRNA splicing and editing, as well as coding single nucleotide polymorphisms, have been associated with numerous cancers, neurological and psychiatirc disorders. With years of experience in Fluorescence in situ hybridization, Creative Bioarray can offer a highly sensitive single-probe FISH/ISH approach that targets short exon/exon RNA splice junction using single-pair oligonucleotide probes. The detection we provide would ensure accurate detection of the mature mRNA variant.
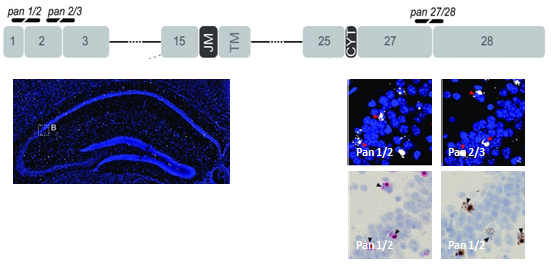 Figure 1. Mouse ErbB4 isoform detection. The specificity and sensitivity of single-pair probes targeting exon-exon boundries were determined by hybridizing sections from WT mice. Probes targeting the exon 1/2 (pan1/2) and exon 2/3 (pan 2/3).
Figure 1. Mouse ErbB4 isoform detection. The specificity and sensitivity of single-pair probes targeting exon-exon boundries were determined by hybridizing sections from WT mice. Probes targeting the exon 1/2 (pan1/2) and exon 2/3 (pan 2/3).
Application
- Sensitive and specific detection of transcripts at cellular level
- Detection of cell-type-specific distribution of isoforms
Features:
- Accurate- In situ Detection Service – Custom design your probe
- Value- We focus on the quality of our service and all supported by competitive pricing
- Efficiency- We are able to provide the fastest turnaround time of any supplier in the industry
Creative Bioarray offers custom Splice Variants Analysis (FISH) services for your scientific research as follows:
- Probe design
- Probe synthesis
- Sample (Cells/FFPE/Others) preparation
- Imaging
- Data analysis
Quotation and ordering
Our customer service representatives are available 24hr a day! We thank you for choosing Creative Bioarray at your preferred Splice Variants Analysis (FISH) services.
References
- Vudattu N K.; et al. Expression analysis and functional activity of interleukin-7 splice variants[J]. Genes and immunity, 2009, 10(2): 132.
- Afsari B.; et al. Splice Expression Variation Analysis (SEVA) for inter-tumor heterogeneity of gene isoform usage in cancer[J]. Bioinformatics, 2018, 34(11): 1859-1867.
- Erben L.; et al. A Novel Ultrasensitive In Situ Hybridization Approach to Detect Short Sequences and Splice Variants with Cellular Resolution[J]. Molecular neurobiology, 2018, 55(7): 6169-6181.
- Deng R.; et al. Highly specific imaging of mRNA in single cells by target RNA-initiated rolling circle amplification[J]. Chemical science, 2017, 8(5): 3668-3675.
- Brinkman B M N. Splice variants as cancer biomarkers[J]. Clinical biochemistry, 2004, 37(7): 584-594.
Explore Other Options
For research use only. Not for any other purpose.

