Kidney Tumor Cells
- Background
- Applications
- Scientific Data
- FAQ
Kidney tumor cells are abnormal cells that proliferate uncontrollably within the kidney, leading to the formation of a mass or tumor. These tumors can be benign (non-cancerous) or malignant (cancerous). The most common type of malignant kidney tumor is renal cell carcinoma (RCC), which originates in the lining of the renal tubules. Kidney tumor cells are often studied in research to understand the disease biology, find new treatment options, and evaluate the effectiveness of drugs. They can be used to create models such as organoids for these purposes.
Types of Kidney Tumor Cells
RCC, the most common form of kidney cancer, can be further subdivided into clear cell, papillary, and chromophobe subtypes, each exhibiting unique morphological and genetic features. Transitional cell carcinoma, originating from the urothelial lining of the renal pelvis, represents another significant category, while Wilms' tumor, a rare pediatric malignancy, also holds a prominent place in the spectrum of kidney tumor cell types.
- Clear cell RCC. This is the most common subtype of RCC, characterized by the presence of clear, glycogen-rich cytoplasm within the tumor cells. The clear cell phenotype is often associated with alterations in the von Hippel-Lindau (VHL) tumor suppressor gene, a key driver of angiogenesis and metabolic reprogramming in these tumor cells.
- Papillary RCC. It is defined by the presence of papillary or tubulopapillary architectural patterns within the tumor. These tumor cells typically exhibit eosinophilic or basophilic cytoplasm and can be further classified into type 1 and type 2 subtypes based on their morphological and genetic characteristics.
- Chromophobe RCC. It is characterized by its distinctive appearance, with tumor cells exhibiting a pale, reticulated cytoplasm and prominent cell borders. This subtype is often associated with genetic alterations in the TP53 and PTEN tumor suppressor genes.
- Wilms' tumor cells. It also called nephroblastoma, is a rare pediatric malignancy that arises from embryonic kidney cells. These tumor cells are characterized by their undifferentiated, blastemal appearance and can exhibit variable histological patterns, including epithelial, stromal, and blastemal components.
Uncovering Molecular Mechanisms
Kidney tumor cells serve as powerful in vitro models for elucidating the molecular underpinnings of cancer initiation, progression, and metastasis. By leveraging advanced techniques, such as genomic profiling, transcriptomics, and proteomics, researchers have delved deep into the genetic and epigenetic alterations that drive the distinctive phenotypes of these tumor cells.
Preclinical Drug Screening and Development
Kidney tumor cell models have become indispensable tools in the preclinical evaluation of novel drug candidates and therapeutic approaches. By employing advanced in vitro assays, such as cell viability, apoptosis, and migration assays, we can rapidly screen and prioritize drug candidates that selectively target the unique vulnerabilities of kidney tumor cells. Researchers leverage these cellular systems to assess the efficacy, selectivity, and safety of potential therapeutic agents, allowing for the identification of promising lead compounds and the optimization of drug formulations.
Elucidating Tumor Microenvironment Interactions
Kidney tumor cells do not exist in isolation; they are deeply embedded within a complex tumor microenvironment that plays a crucial role in their behavior and therapeutic response. By studying the dynamic interplay between kidney tumor cells and their microenvironment, researchers have gained valuable insights into the mechanisms of immune evasion, angiogenesis, and metastasis.
LZTFL1 Inhibits ccRCC Cell Growth and Proliferation in Vitro and in Vivo
LZTFL1 is a tumor suppressor located in chromosomal region 3p21.3 that is deleted frequently and early in various cancer types including kidney cancer. To investigate the pathophysiological role of LZTFL1 in clear cell RCC (ccRCC), we analyzed LZTFL1 expression in various established ccRCC cell lines. Among eight ccRCC cell lines we tested, LZTFL1 expression is downregulated in ACHN, Caki1, and RCCJF (Fig. 1a). We re-expressed LZTFL1 stably in low-LZTFL1 expressing ACHN and Caik1 cell lines and knocked down LZTFL1 in high-LZTFL1 expressing A498 cell line (Fig. 1b). Proliferation assays by Cell Counting Kit-8 (CCK-8) and colony formation assays revealed that over-expression of LZTFL1 in ACHN and Caki1 cells inhibited cell growth and proliferation capacity. Conversely, the knockdown of LZTFL1 in A498 cells enhanced cell growth and colony formation ability significantly (Fig. 1c, d). Overexpression of LZTFL1 also reduced the size and weight of subcutaneous xenografts in vivo (Fig. 1e). Conversely, knockdown of LZTFL1 promoted the growth of A498 xenograft in vivo (Fig. 1f).
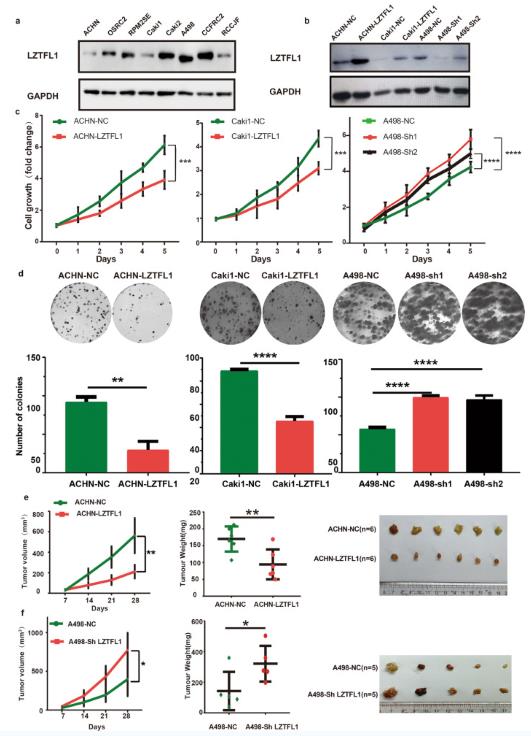 Fig.
1 LZTFL1 suppresses cell proliferation in vitro and in vivo. (Lu J, et al, 2023)
Fig.
1 LZTFL1 suppresses cell proliferation in vitro and in vivo. (Lu J, et al, 2023)
VHLOverexpression Alters Protein Expression Profiling in ccRCC Cells
Quantitative proteomics analysis was conducted on VHL-OE and control cells in biological triplicates for identifying differentially expressed proteins (DEPs). We identified 9459 proteins and 7037 proteins separately with two and more unique peptides quantified. The quantification ratios of biological replicates showed a high correlation, indicating that the results were reproducible (R2 = 0.9) (Fig. 3A). The quantitative ratios were filtered by population statistics to calculate the threshold cut-off as a 50% variation for DEPs. Volcano diagram showed that 404 proteins were up-regulated [VHL-OE/Control > 1.5; false discovery rate (FDR) adjusted P value < 0.05], and 228 proteins were down-regulated (VHL-OE/Control < 0.67; FDR adjusted P value < 0.05) (Fig. 2B). VHL expression was significantly up-regulated, and its substrate HIF1α was significantly down-regulated (Fig. 2B). The expression levels of proteins involved in glycolysis including Glyceraldehyde-3-phosphate dehydrogenase (GAPDH), Triosephosphate isomerase (TPI1), Enolase 2 (ENO2), Pyruvate kinase PKLR (PKLR), and Pyruvate kinase PKM (PKM) were down-regulated in VHL-OE cells compared with those in control cells (Fig. 2C), whereas the expression levels of many subunits involved in the oxidative phosphorylation pathway, including subunits of Complex I, Complex II, Complex III, and Complex IV, were up-regulated in VHL-OE cells (Fig. 2C). The changes of GLUT1, ENO2, and HK2 were validated by RT-PCR (Fig. 2D), while the changes of GAPDH, NDUFV2, and NDUFA4 were further validated by Western blotting (Fig. 2E).
The top 20 pathways enriched by IPA revealed that VHL regulated four major biological processes including metabolism, immune regulation, apoptosis, and cell movement (Fig. 2F). The proteins involved in oxidative phosphorylation, fatty acid oxidation, mitochondrial L-carnitine shuttling, cholesterol biosynthesis, mevalonate pathway, and geranylgeranyl-diphosphate biosynthesis were mostly up-regulated. The number of up-regulated proteins involved in apoptosis and cell movement-related pathways was more than that of the down-regulated proteins (Fig. 2F). Furthermore, we calculated the Z-scores of enriched biological pathways by using the core analysis method in IPA, which assesses the match of observed and predicted up- or down-regulation patterns and represents the non-randomness of directionality within a gene set. The results showed that the apoptotic pathways had Z-scores above 2, suggesting that apoptotic pathways were activated after VHL overexpression. On the other hand, the cell motility-related pathways had Z-scores below −2, suggesting that cell movement-related pathways were inhibited after VHL overexpression. These results indicated that VHL overexpression reduced cell viability and motility.
Proteomics analysis also identified that major histocompatibility antigens HLA-A, HLA-B, HLA-C, HLA-E, and HLA-H were up-regulated in VHL-OE cells (Fig. 2B). The changes were further validated by RT-PCR analysis, showing that the mRNA levels of HLA-A, HLA-B, and HLA-E were also up-regulated (Fig. 2G). This was confirmed by Western blotting, in which both MHC class I molecule HLA-A and MHC class II molecule HLA-DR were up-regulated (3.2-fold and 4.5-fold higher in VHL-OE cells than in control cells, respectively) (Fig. 2H). Intriguingly, PD-L1, an immune checkpoint protein, was down-regulated in VHL-OE cells at both mRNA and protein levels (Fig. 2G and H). Consistently, the gene set enrichment analysis (GSEA) revealed that the expression of antigen processing and presentation signature genes was increased in VHL-OE cells compared with that in control cells (Fig. 2I). These results suggest that VHL overexpression up-regulates both MHC class I and class II molecules, thus enhancing antigen processing and presentation.
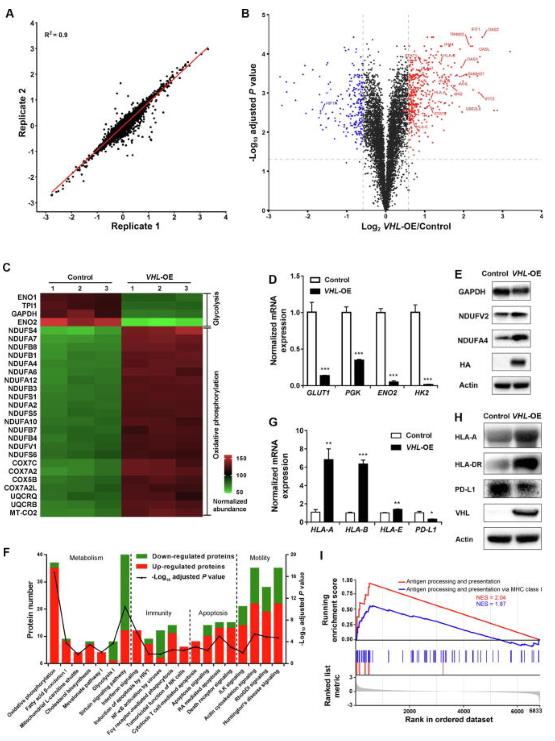 Fig.
2 Stable Knockdown of HRP-3 in HCC cells sensitized cells to become shrunk morphology of apoptosis under the
energy pressure. (Cai H, et al., 2015)
Fig.
2 Stable Knockdown of HRP-3 in HCC cells sensitized cells to become shrunk morphology of apoptosis under the
energy pressure. (Cai H, et al., 2015)
RCC is the most common type of kidney cancer. It starts in the lining of the small tubes in the kidney and can grow into a mass or tumor.
Kidney tumor cells are used to study the disease biology of kidney cancer, including how it develops and progresses. They are also used to test the effectiveness of various drugs and to develop new therapies.
Kidney tumor cell lines allow for rapid and cost-effective screening of large chemical libraries to identify promising drug candidates. These cellular systems enable the evaluation of drug efficacy, selectivity, and safety, facilitating the optimization of lead compounds before advancing to animal studies and clinical trials.
Filters Clear all filters
Species
- African clawed frog (1)
- American mink (1)
- Asian tiger mosquito (1)
- Atlantic salmon (1)
- Bluegill (2)
- Bluestriped grunt (1)
- Bovine (7)
- Brazilian free-tailed bat (1)
- Brown bullhead (2)
- Cabbage looper (1)
- Cabbage moth (6)
- Cat (4)
- Central mudminnow (1)
- Chicken (3)
- Chinese hamster (5)
- Chinook salmon (2)
- Chum salmon (1)
- Coho salmon (1)
- Common carp (2)
- Cotton-top tamarin (1)
- Dog (2)
- Fall armyworm (3)
- Fathead minnow (2)
- Fruit fly (1)
- Gilthead sea bream (2)
- Golden hamster (7)
- Goldfish (6)
- Gray dwarf hamster (1)
- Green monkey (2)
- Gypsy moth (1)
- Horse (1)
- Human (999)
- Japanese eel (1)
- Japanese rice fish (7)
- Koi carp (1)
- Mouse (315)
- Mouse x Gray dwarf hamster (1)
- Mouse x Rat (20)
- Northern pike (1)
- Pig (3)
- Rabbit (2)
- Rainbow trout (3)
- Rat (115)
- Rhesus macaque (1)
- Salt marsh moth (1)
- Sheep (2)
- Snakehead murrel (2)
- Sockeye salmon (1)
- Vervet monkey (2)
- Zebrafish (2)
Source
- Abdomen (1)
- Abdomen Metastasis (2)
- Adipose (2)
- Adrenal Gland (8)
- Adrenal Gland Metastasis (2)
- Aorta (4)
- Artery (1)
- Ascites (28)
- Ascites Metastasis (37)
- Bile Duct (3)
- Bladder (25)
- Bladder Metastasis (1)
- Blastocyst (1)
- Blastula (1)
- Blood (127)
- Bone (27)
- Bone Marrow (57)
- Bone Marrow Metastasis (18)
- Bone Metastasis (6)
- Brain (55)
- Brain Metastasis (8)
- Breast (30)
- Bronchus (1)
- Caudal Peduncle (1)
- Caudal Trunk (2)
- Cecum (3)
- Cerebrospinal Fluid (1)
- Cerebrospinal Fluid Metastasis (1)
- Cervix (32)
- Colon (90)
- Connective Tissue (7)
- Cornea (3)
- Cutaneous Metastasis (1)
- Dermis (2)
- Duodenum (1)
- Embryo (29)
- Endometrium (17)
- Esophagus (44)
- Eye (12)
- Eye Socket (5)
- Fetus (3)
- Fin (9)
- Foreskin (4)
- Gallbladder (1)
- Gingiva (2)
- Globe (2)
- Glomerulus (2)
- Groin (1)
- Head Kidney (2)
- Heart (4)
- Hemolymph (1)
- Hypodermis Metastasis (5)
- Ileum (1)
- Intestine (94)
- Jejunum (1)
- kidney (1)
- Kidney (27)
- Liver (35)
- Liver Metastasis (17)
- Lung (58)
- Lung Metastasis (8)
- Lymph Node (8)
- Lymph Node Metastasis (59)
- Muscle (7)
- Muscle Metastasis (2)
- Nose (2)
- Omentum Metastasis (2)
- Oral Cavity (10)
- Ovary (21)
- Ovary Metastasis (2)
- Pancreas (19)
- Pelvic Wall Metastasis (1)
- Pelvis (1)
- Perianal Space Metastasis (1)
- Pericardial Effusion (1)
- Pericardial Effusion Metastasis (2)
- Perineus (1)
- Peripheral Blood (126)
- Peripheral Nervous System (21)
- Peritoneal Effusion (2)
- Peritoneum (1)
- Peritoneum Metastasis (1)
- Pharynx (3)
- Pituitary Gland (7)
- Pleural Effusion (54)
- Pleural Effusion Metastasis (46)
- Prostate (7)
- Rectum (15)
- Renal Pelvis (1)
- Retroperitoneal Space (2)
- Salivary Gland (2)
- Skeletal Muscle (5)
- Skin (32)
- Skin Metastasis (3)
- Small Intestine (4)
- Small Intestine Metastasis (1)
- Smooth Muscle (2)
- Soft Tissue (1)
- Soft Tissue Metastasis (1)
- Spinal Cord (2)
- Stomach (4)
- Testis (15)
- Thoracic Cavity Metastasis (6)
- Thymus (5)
- Thyroid Gland (16)
- Thyroid Gland Metastasis (1)
- Tongue (5)
- Trachea (1)
- Umbilical Cord (1)
- Umbilical Cord Blood (1)
- Urachus (1)
- Ureter (1)
- Uterus (54)
- Uvea (2)
- Vagina (2)
- Vulva (1)
Disease
- Acute Biphenotypic Leukemia (1)
- Acute Erythroid Leukemia (4)
- Acute Megakaryoblastic Leukemia (4)
- Acute Monocytic Leukemia (9)
- Acute Myeloid Leukemia (25)
- Acute Promyelocytic Leukemia (2)
- Adrenal Gland Neuroblastoma (11)
- Adult B Acute Lymphoblastic leukemia (1)
- Adult B Acute Lymphoblastic Leukemia (6)
- Adult T Acute Lymphoblastic Leukemia (6)
- Adult T Lymphoblastic Lymphoma (2)
- Adult T-Cell Leukemia/Lymphoma (1)
- Alveolar Rhabdomyosarcoma (4)
- Alveolar Ridge Squamous Cell Carcinoma (1)
- Amelanotic Melanoma (3)
- Ampulla of Vater Adenocarcinoma (1)
- Ampulla of Vater Adenosquamous Carcinoma (3)
- Anaplastic Astrocytoma (3)
- Anaplastic Large Cell Lymphoma (7)
- Askin Tumor (1)
- Astrocytoma (5)
- B Acute Lymphoblastic Leukemia (2)
- B-Cell Non-Hodgkin Lymphoma (5)
- Bare Lymphocyte Syndrome Type 2 (1)
- Barrett Adenocarcinoma (2)
- Benign Prostatic Hyperplasia (1)
- Bladder Carcinoma (14)
- Bladder Squamous Cell Carcinoma (1)
- Bovine Leukemia (2)
- Breast Adenocarcinoma (4)
- Breast Carcinoma (9)
- Breast Ductal Carcinoma (2)
- Burkitt Lymphoma (17)
- Canavan Disease (1)
- Canine Histiocytic Sarcoma (1)
- Cecum Adenocarcinoma (3)
- Central Nervous System Lymphoma (2)
- Cervical Adenocarcinoma (2)
- Cervical Adenosquamous Carcinoma (2)
- Cervical Small Cell Carcinoma (1)
- Cervical Squamous Cell Carcinoma (2)
- Chicken Bursal Lymphoma (2)
- Childhood B Acute Lymphoblastic Leukemia (13)
- Childhood T Acute Lymphoblastic Leukemia (16)
- Childhood T Lymphoblastic Lymphoma (1)
- Cholangiocarcinoma (2)
- Chronic Eosinophilic Leukemia (1)
- Chronic Lymphocytic Leukemia (2)
- Chronic Myeloid Leukemia (23)
- Clear Cell Renal Cell Carcinoma (2)
- Colon Adenocarcinoma (55)
- Colon Carcinoma (34)
- Colorectal Adenocarcinoma (1)
- Colorectal Carcinoma (1)
- Congenital Pure Red Cell Aplasia (1)
- Cutaneous Melanoma (10)
- Dedifferentiated Chondrosarcoma (1)
- Desmoplastic Melanoma (1)
- Diffuse Large B-Cell Lymphoma (29)
- Down Syndrome (2)
- EBV-Related Burkitt Lymphoma (12)
- Embryonal Carcinoma (3)
- Embryonal Rhabdomyosarcoma (3)
- Endometrial Adenocarcinoma (13)
- Endometrial Adenosquamous Carcinoma (2)
- Endometrial Carcinoma (2)
- Endometrioid Stromal Sarcoma (1)
- Epithelioid Hemangioendothelioma (1)
- Epithelioid Sarcoma (3)
- Esophageal Adenocarcinoma (6)
- Esophageal Squamous Cell Carcinoma (41)
- Essential Thrombocythemia (1)
- Ewing Sarcoma (2)
- Extraskeletal Myxoid Chondrosarcoma (1)
- Fanconi Anemia (1)
- Fibrosarcoma (1)
- Follicular Lymphoma (2)
- Gallbladder Carcinoma (2)
- Gallbladder Undifferentiated Carcinoma (2)
- Gastric Adenocarcinoma (6)
- Gastric Adenosquamous Carcinoma (1)
- Gastric Carcinoma (5)
- Gastric Choriocarcinoma (1)
- Gastric Fundus Carcinoma (1)
- Gastric Signet Ring Cell Adenocarcinoma (1)
- Gastric Small Cell Carcinoma (2)
- Gastric Tubular Adenocarcinoma (5)
- Gastroesophageal Junction Adenocarcinoma (1)
- Gestational Choriocarcinoma (1)
- Gingival Squamous Cell Carcinoma (2)
- Glioblastoma (18)
- Gliosarcoma (1)
- Goldfish Erythrophoroma (4)
- Hairy Cell Leukemia (1)
- Hamster Kidney Tumor (1)
- Hamster Pancreatic Ductal Adenocarcinoma (1)
- Hamster Uterine Leiomyosarcoma (1)
- Hepatoblastoma (2)
- Hepatocellular Carcinoma (6)
- Hepatosplenic T-Cell Lymphoma (2)
- Hereditary Thyroid Gland Medullary Carcinoma (1)
- High Grade B-Cell Lymphoma (1)
- High Grade Ovarian Serous Adenocarcinoma (8)
- Hodgkin Lymphoma (9)
- Hypopharyngeal Squamous Cell Carcinoma (2)
- Infectious Mononucleosis (1)
- Intrahepatic Cholangiocarcinoma (6)
- Invasive Breast Carcinoma of No Special Type (12)
- Invasive Breast Lobular Carcinoma (1)
- Kidney Neoplasm (1)
- Kidney Rhabdoid Tumor (1)
- Krukenberg Tumor (1)
- Liposarcoma (1)
- Lung Adenocarcinoma (17)
- Lung Giant Cell Carcinoma (8)
- Lung Large Cell Carcinoma (9)
- Lung Mucoepidermoid Carcinoma (1)
- Lung Non-Small Cell Carcinoma (2)
- Lung Small Cell Carcinoma (25)
- Lung Squamous Cell Carcinoma (9)
- Lymphoblastic Lymphoma (1)
- Malignant Peripheral Nerve Sheath Tumor (1)
- Mantle Cell Lymphoma (5)
- Mature Gastric Teratoma (1)
- Maxillary Sinus Squamous Cell Carcinoma (1)
- Medaka Hepatoma (2)
- Medulloblastoma (3)
- Melanoma (24)
- Meningioma (2)
- Minimally Invasive Lung Adenocarcinoma (1)
- Monophasic Synovial Sarcoma (1)
- Mouse Bladder Transitional Cell Carcinoma (1)
- Mouse Chondrosarcoma (1)
- Mouse Colon Adenocarcinoma (3)
- Mouse Ependymoma (2)
- Mouse Erythroid Leukemia (13)
- Mouse Fibrosarcoma (5)
- Mouse Glioblastoma (1)
- Mouse Hemangioendothelioma (1)
- Mouse Hepatocellular Carcinoma (1)
- Mouse Insulinoma (3)
- Mouse Intestinal Tract Neuroendocrine Adenoma (1)
- Mouse Islet Cell Adenoma (1)
- Mouse Kidney Carcinoma (1)
- Mouse Leukemia (10)
- Mouse Leydig Cell Tumor (1)
- Mouse Lymphoma (8)
- Mouse Mammary Gland Malignant Neoplasm (23)
- Mouse Melanoma (9)
- Mouse Multiple Myeloma (5)
- Mouse Myeloid Leukemia (3)
- Mouse Neoplasm (1)
- Mouse Neuroblastoma (21)
- Mouse Oral Cavity Squamous Cell Carcinoma (1)
- Mouse Osteosarcoma (3)
- Mouse Pituitary Gland Neoplasm (1)
- Mouse Plasmacytoma (1)
- Mouse Precursor T Cell Lymphoblastic Lymphoma/Leukemia (2)
- Mouse Pulmonary Adenoma (1)
- Mouse Pulmonary Malignant Tumor (3)
- Mouse Pulmonary Squamous Cell Carcinoma (1)
- Mouse Rectum Carcinoma (2)
- Mouse Reticulum Cell Sarcoma (2)
- Mouse Sarcoma (1)
- Mouse Teratocarcinoma (8)
- Mouse Thymic Lymphoma (3)
- Mycosis Fungoides (1)
- Myelodysplastic Syndrome (1)
- Myxofibrosarcoma (1)
- Natural Killer Cell Lymphoblastic Leukemia/Lymphoma (2)
- Neuroblastoma (26)
- Oral Cavity Squamous Cell Carcinoma (15)
- Osteoid Osteoma (1)
- Osteosarcoma (15)
- Ovarian Carcinoma (1)
- Ovarian Clear Cell Adenocarcinoma (1)
- Ovarian Endometrioid Adenocarcinoma (4)
- Ovarian Granulosa Cell Tumor (1)
- Ovarian Mucinous Adenocarcinoma (2)
- Ovarian Serous Adenocarcinoma (2)
- Ovarian Serous Cystadenocarcinoma (2)
- Ovarian Small Cell Carcinoma (1)
- Pancreatic Adenocarcinoma (13)
- Pancreatic Carcinoma (5)
- Pancreatic Ductal Adenocarcinoma (12)
- Papillomavirus-Independent Cervical Squamous Cell Carcinoma (1)
- Papillomavirus-Related Cervical Adenocarcinoma (7)
- Papillomavirus-Related Cervical Squamous Cell Carcinoma (4)
- Papillomavirus-Related Endocervical Adenocarcinoma (16)
- Paroxysmal Nocturnal Hemoglobinuria (3)
- Pharyngeal Squamous Cell Carcinoma (1)
- Plasma Cell Myeloma (15)
- Pleural Epithelioid Mesothelioma (5)
- Pleural Sarcomatoid Mesothelioma (2)
- Poorly Differentiated Thyroid Gland Carcinoma (1)
- Primary Cutaneous T-Cell Non-Hodgkin Lymphoma (1)
- Primary Effusion Lymphoma (7)
- Primitive Neuroectodermal Tumor (1)
- Prostate carcinoma (1)
- Prostate Carcinoma (9)
- Rat C-Cell Carcinoma (1)
- Rat Cholangiocarcinoma (1)
- Rat Colon Adenocarcinoma (5)
- Rat Digestive System Neoplasm (1)
- Rat Fibrosarcoma (1)
- Rat Hepatocellular Carcinoma (20)
- Rat Histiocytic Sarcoma (1)
- Rat Insulinoma (2)
- Rat Leukemia (1)
- Rat Leydig Cell Adenoma (1)
- Rat Lung Carcinoma (1)
- Rat Malignant Glioma (4)
- Rat Malignant Meningioma (1)
- Rat Malignant Oligodendroglioma (2)
- Rat Malignant Thymoma (3)
- Rat Mammary Gland Adenocarcinoma (10)
- Rat Neuroblastoma (3)
- Rat Osteosarcoma (2)
- Rat Pituitary Gland Neoplasm (6)
- Rat Prostate Adenocarcinoma (3)
- Rat Rhabdomyosarcoma (1)
- Rat Sarcoma (2)
- Rat Squamous Cell Carcinoma (1)
- Rat Urinary Bladder Transitional Cell Carcinoma (2)
- Rat Urinary System Neoplasm (6)
- Rectal Adenocarcinoma (13)
- Rectosigmoid Adenocarcinoma (1)
- Recurrent Bladder Carcinoma (1)
- Renal Cell Carcinoma (7)
- Renal Pelvis Urothelial Carcinoma (1)
- Retinoblastoma (11)
- Sacral Chordoma (1)
- Sacrococcygeal Teratoma (1)
- Salivary Gland Squamous Cell Carcinoma (1)
- Sezary Syndrome (1)
- Shwachman-Diamond Syndrome (1)
- Skin Squamous Cell Carcinoma (2)
- Splenic Marginal Zone Lymphoma (1)
- Testicular Embryonal Carcinoma (8)
- Testicular Teratoma (2)
- Testicular Yolk Sac Tumor (1)
- Thyroid Gland Anaplastic Carcinoma (10)
- Thyroid Gland Follicular Carcinoma (4)
- Thyroid Gland Papillary Carcinoma (3)
- Thyroid Gland Sarcoma (1)
- Thyroid Gland Squamous Cell Carcinoma (2)
- Tongue Adenosquamous Carcinoma (1)
- Tongue Squamous Cell Carcinoma (6)
- Type I Endometrial Adenocarcinoma (1)
- Ureter Urothelial Carcinoma (1)
- Uterine Carcinosarcoma (2)
- Uterine Corpus Leiomyosarcoma (1)
- Uterine Corpus Sarcoma (2)
- Uveal Melanoma (2)
- Vaginal Melanoma (2)
- Vulvar Melanoma (1)
- Vulvar Squamous Cell Carcinoma (1)
Description: The cells express the transforming gene of adenovirus 5. Adenovirus 5 DNA from both the right and ...
Description: Established from the malignant pleural effusion of a 75-year-old man with kidney carcinoma in 1987
Description: Established from the sarcomatoid component of a grade III transitional cell carcinoma of the renal ...
Description: Human cell line derived from neuroblastoma of adrenal gland.
Description: Human cell line derived from neuroblastoma.
Description: Japanese renal carcinoma cells expressing HLA-A2402. Cell growth is slow.
Description: Japanese renal carcinoma cells. Cell growth is slow.
Description: Japanese kidney carcinoma cells. Cell growth is slow.
Description: Kidney carcinoma from a Japanese patient. Cell growth is slow.
Description: Rhabdoid tumor of kidney (formerly classified as Wilms' tumor)
Description: Renal tumor cell from a Japanese. Transplantable to nude mice.
Description: Species: human - female, 9 months old, CaucasianIsoenzyme: G6PD, BKaryology: diploidHistopathology: ...
Description: OACP4 C was established from a primary tumour located in gastric cardia of a 55 year-old Caucasian ...