Spatial Proteomics Solutions
- Service Details
- Features
- Applications
- FAQ
- Explore Other Options
From Cell to Space: Comprehensive Spatial Proteomics Solutions Await
Cellular heterogeneity refers to the significant functional and structural differences that exist among cells that appear identical on the outside. Traditional studies often rely on signal averages from entire tissues or large cell populations, lacking spatial information about individual cells, which prevents scientists from fully understanding complex biological systems. Spatial proteomics technology addresses this challenge by providing a detailed map of protein expression across different regions and cell types within tissues. Combining protein data with spatial information helps us better understand tissue environments to find new biomarkers and understand biological functions.
Creative Bioarray's spatial proteomics services primarily utilize antibody-based fluorescence imaging and LCM-MS techniques. LCM-MS stands out for its advanced accuracy which lets researchers take precise tissue parts and then test mass spectrometry data to show protein expression changes between areas. Our technology shows protein locations precisely and tracks their movement within cells to help researchers better understand cellular processes and diseases. At Creative Bioarray, we are committed to advancing scientific discovery through cutting-edge technology, empowering researchers to achieve breakthrough progress.
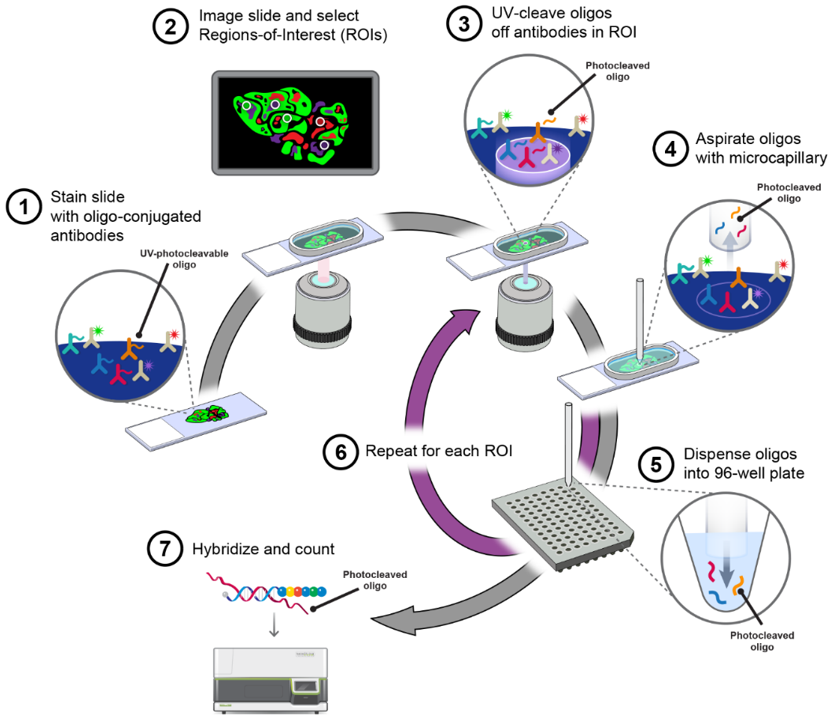
 Examples of related findings and troubleshooting in spatial proteomics (Wu M, Tao H, et al., 2024).
Examples of related findings and troubleshooting in spatial proteomics (Wu M, Tao H, et al., 2024).
Our Spatial Proteomics Technology
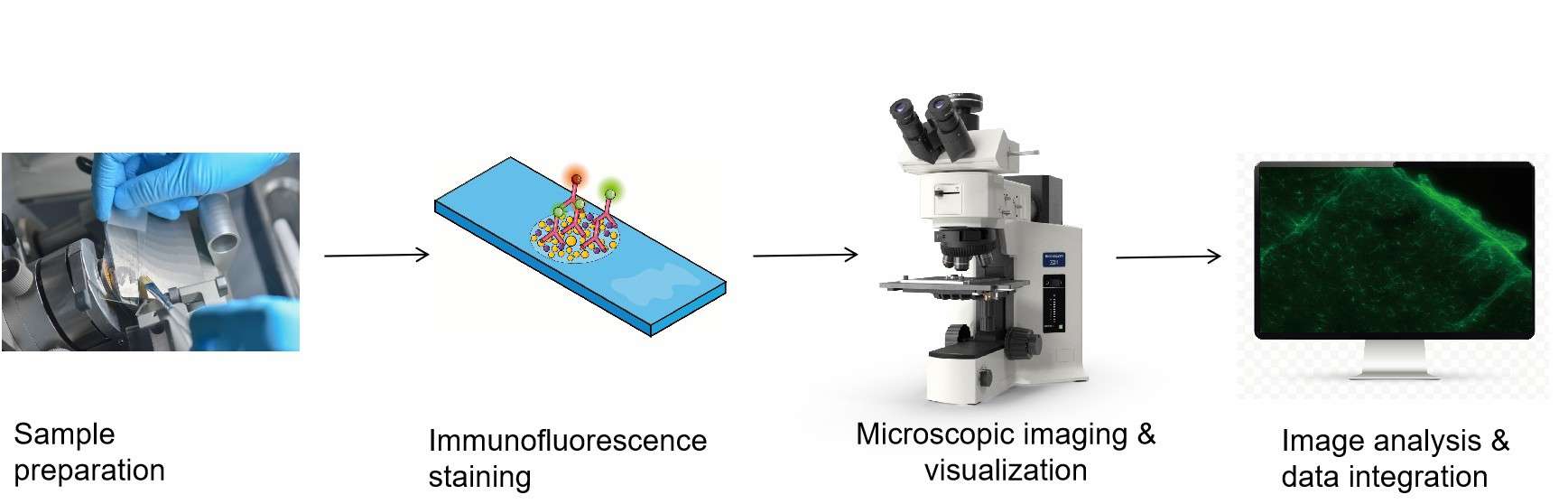
Workflow
1
Sample preparation
Pre-treatment is performed based on the sample type (such as paraffin-embedded tissue or frozen tissue), including steps like deparaffinization, hydration, and antigen retrieval. A high-precision microtome is used to section the processed samples into thin slices, typically 4-6 micrometers thick, which are then mounted onto adhesion slides.
2
Immunofluorescence staining
Utilizes cyclic immunofluorescence staining technology where antibodies are used in conjunction with continuous elution processes to enable high-throughput protein detection.
3
High-Resolution imaging
Employing laser scanning confocal microscopy, complemented by automated high-definition tissue imaging systems, to scan each fluorescence channel.
4
Tissue damage control and quality monitoring
H&E images are used for quality control of slice integrity, ensuring the reliability of the experiment.
5
Image analysis and data integration
The captured images undergo quantitative analysis, assessing protein expression levels and spatial distribution features. Data from each staining round are integrated to construct protein co-localization maps, revealing interactions among proteins.
Technical Advantages
High-Resolution Imaging
Multiplexing Capabilities
Non-destructive
Workflow
1
Tissue sectioning and staining
Select fresh or fixed tissue sections (such as FFPE) for H&E staining or immunofluorescence staining to mark the target areas.
2
Laser capture microdissection
Under a microscope, circle the target area, use a laser to cut it out, and collect the tissue fragments into collection tubes.
3
Protein extraction and digestion
Extract proteins and perform enzymatic digestion to convert proteins into peptides, preparing them for mass spectrometry analysis.
4
Mass spectrometry analysis
Use high-resolution mass spectrometers to detect, identify, and quantify proteins. Bioinformatics analysis follows.
5
Data analysis
Employ image analysis software to correlate mass spectrometry data with the spatial information of the slices, perform baseline correction and peak fitting, and conduct in-depth differential analysis using data visualization tools.
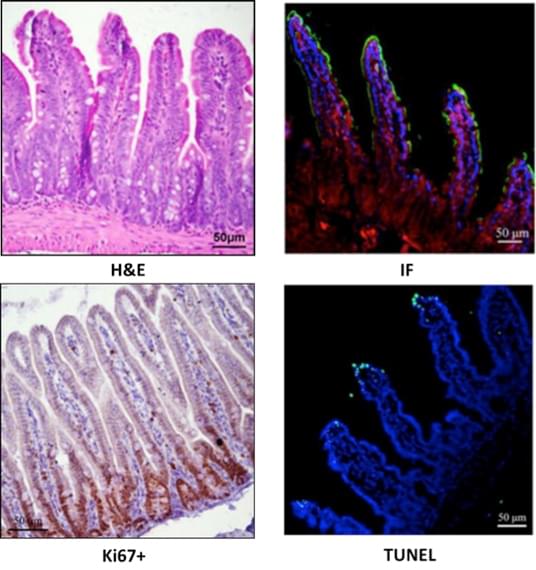
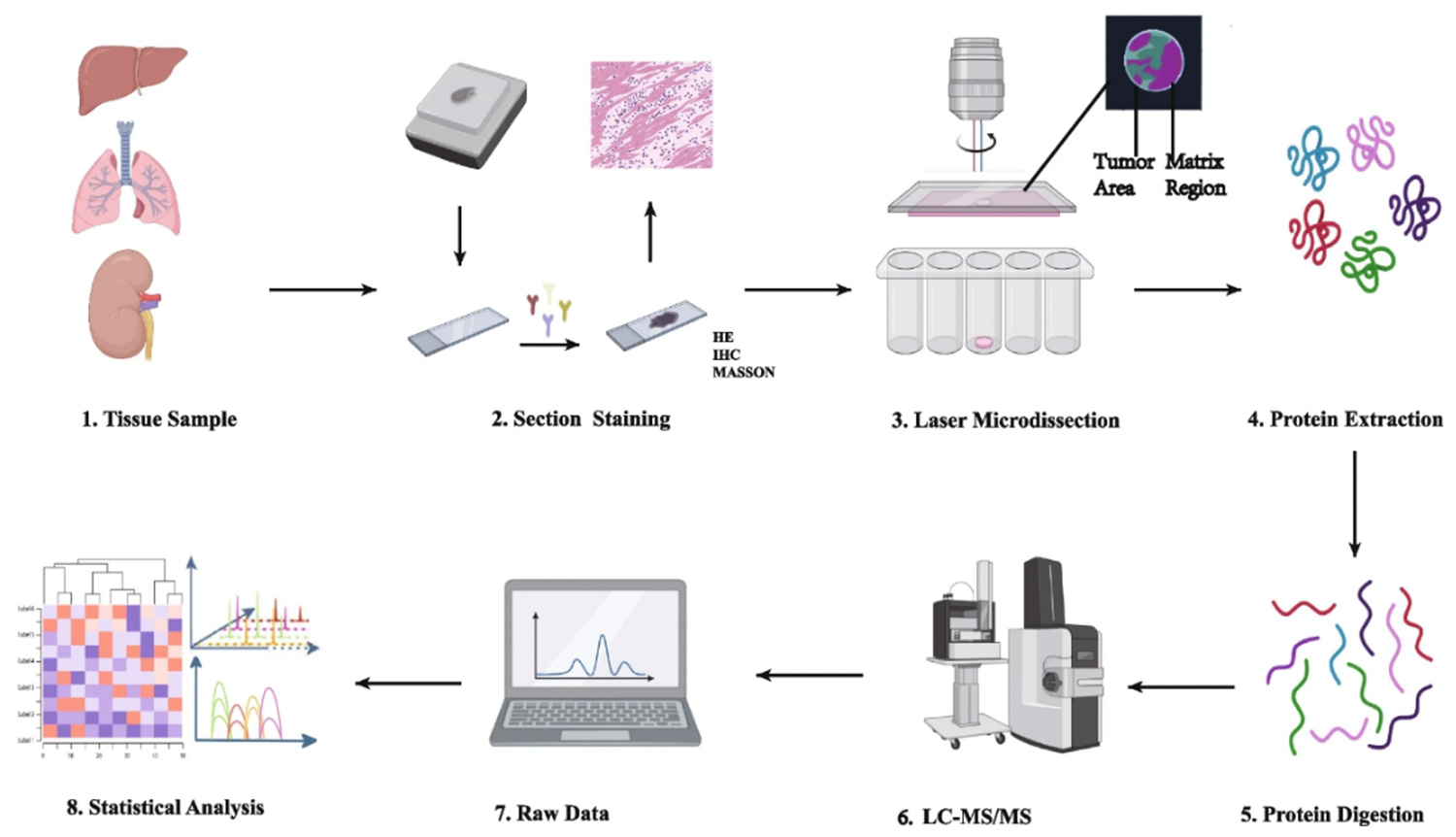 Operational procedure of spatial proteomics mass spectrometry methods (Wu M, Tao H, et al., 2024).
Operational procedure of spatial proteomics mass spectrometry methods (Wu M, Tao H, et al., 2024).
Technical Advantages
Meaning unbiased
Deeper coverage of molecules
High-sensitivity detection
Features
Full-Service Workflow
From sample preparation to data analysis, we offer a one-stop solution that simplifies the experimental process for our clients.
Extensive Project Experience
Our wealth of practical experience ensures that we deliver high-quality services to our clients.
High-Throughput and In-Depth Analysis
We provide high-throughput, in-depth proteomic analysis at both single-cell and subcellular levels.
Customized Solutions
We design personalized experiments tailored to the client's research needs, ensuring alignment with experimental goals and focus areas.
Multi-Omics Integration
By integrating spatial transcriptomics, metabolomics, and other omics data, we deliver more comprehensive biological insights.
What Can You Do with Our Spatial Proteomics Solutions?
- Tumor research: Study of tumor heterogeneity and development, as well as changes in the tumor microenvironment under different treatments.
- Neuroscience research: Analyze subcellular localization of proteins in neural tissue and their functions.
- Immune cell infiltration research: Examine the protein networks of immune cells within tissues, along with infiltration patterns and interactions.
- Disease biomarker discovery: Identify protein expression patterns related to diseases, uncovering new biomarkers.
- Spatial single-cell analysis: Conduct spatial distance measurements, cell neighborhood and phenotypic feature analysis to discover spatial characteristics of rare cell types and functional states.
FAQ
1. What is the difference between spatial proteomics and traditional proteomics?
The main distinction between spatial proteomics and regular proteomics lies in spatial proteomics' capacity to show protein positions inside tissues. Standard proteomics studies measure protein types and amounts but does not track their exact tissue positions. Spatial proteomics merges protein location details with biological data to show how proteins work with tissue structure and cell neighbors which helps scientists understand complex tissues.
2. What are the requirements for samples to be tested?
Researchers can test fresh tissues directly or use frozen sections and paraffin-embedded specimens. Fresh tissue samples need immediate processing after removal and need to stay cold during transport. Frozen tissue sections must remain cold to prevent excessive freezing and thawing. Paraffin-embedded tissue sections should be of good quality, typically with a thickness of 3-5 micrometers. Additionally, samples should be clearly labeled with information about their source and handling.
3. Can LCM-MS technology perform protein quantification?
Yes, LCM-MS technology can perform both qualitative analysis (identification of protein types) and quantitative analysis (measurement of relative or absolute protein abundance). This is typically achieved by comparing the mass spectrometry signal intensities of the same peptide segments across different samples.
4. What are the advantages of LCM-MS technology?
The advantages of LCM-MS technology include its high precision, high throughput, and ability to provide spatial distribution information of proteins within tissues. It combines the high precision sample collection capability of LCM with the high throughput analysis capability of mass spectrometry, allowing scientists to deeply investigate the composition and function of proteins within complex tissue environments.
5. How are the results of spatial proteomics services presented?
Results are typically displayed through visualized spatial maps and data analysis software, showing the spatial distribution and abundance of proteins in tissue sections. Researchers can observe the spatial localization of various protein markers, distribution of cell types, and their interactions. Additionally, data analysis may include detailed statistical reports to assist in the interpretation of complex data.
References
- Wu M, Tao H, et al. Spatial proteomics: unveiling the multidimensional landscape of protein localization in human diseases. Proteome Sci. 2024. 22(1):7.
- Chen T, You L, et al. Spatial Transcriptomic Technologies. Cells. 2023. 12(16):2042.
Related Services
Explore Other Options