- You are here: Home
- Services
- In Vitro DMPK Services
- Drug-Drug Interaction
- CYP and UGT Reaction Phenotyping Assay
Services
-
Cell Services
- Cell Line Authentication
- Cell Surface Marker Validation Service
-
Cell Line Testing and Assays
- Toxicology Assay
- Drug-Resistant Cell Models
- Cell Viability Assays
- Cell Proliferation Assays
- Cell Migration Assays
- Soft Agar Colony Formation Assay Service
- SRB Assay
- Cell Apoptosis Assays
- Cell Cycle Assays
- Cell Angiogenesis Assays
- DNA/RNA Extraction
- Custom Cell & Tissue Lysate Service
- Cellular Phosphorylation Assays
- Stability Testing
- Sterility Testing
- Endotoxin Detection and Removal
- Phagocytosis Assays
- Cell-Based Screening and Profiling Services
- 3D-Based Services
- Custom Cell Services
- Cell-based LNP Evaluation
-
Stem Cell Research
- iPSC Generation
- iPSC Characterization
-
iPSC Differentiation
- Neural Stem Cells Differentiation Service from iPSC
- Astrocyte Differentiation Service from iPSC
- Retinal Pigment Epithelium (RPE) Differentiation Service from iPSC
- Cardiomyocyte Differentiation Service from iPSC
- T Cell, NK Cell Differentiation Service from iPSC
- Hepatocyte Differentiation Service from iPSC
- Beta Cell Differentiation Service from iPSC
- Brain Organoid Differentiation Service from iPSC
- Cardiac Organoid Differentiation Service from iPSC
- Kidney Organoid Differentiation Service from iPSC
- GABAnergic Neuron Differentiation Service from iPSC
- Undifferentiated iPSC Detection
- iPSC Gene Editing
- iPSC Expanding Service
- MSC Services
- Stem Cell Assay Development and Screening
- Cell Immortalization
-
ISH/FISH Services
- In Situ Hybridization (ISH) & RNAscope Service
- Fluorescent In Situ Hybridization
- FISH Probe Design, Synthesis and Testing Service
-
FISH Applications
- Multicolor FISH (M-FISH) Analysis
- Chromosome Analysis of ES and iPS Cells
- RNA FISH in Plant Service
- Mouse Model and PDX Analysis (FISH)
- Cell Transplantation Analysis (FISH)
- In Situ Detection of CAR-T Cells & Oncolytic Viruses
- CAR-T/CAR-NK Target Assessment Service (ISH)
- ImmunoFISH Analysis (FISH+IHC)
- Splice Variant Analysis (FISH)
- Telomere Length Analysis (Q-FISH)
- Telomere Length Analysis (qPCR assay)
- FISH Analysis of Microorganisms
- Neoplasms FISH Analysis
- CARD-FISH for Environmental Microorganisms (FISH)
- FISH Quality Control Services
- QuantiGene Plex Assay
- Circulating Tumor Cell (CTC) FISH
- mtRNA Analysis (FISH)
- In Situ Detection of Chemokines/Cytokines
- In Situ Detection of Virus
- Transgene Mapping (FISH)
- Transgene Mapping (Locus Amplification & Sequencing)
- Stable Cell Line Genetic Stability Testing
- Genetic Stability Testing (Locus Amplification & Sequencing + ddPCR)
- Clonality Analysis Service (FISH)
- Karyotyping (G-banded) Service
- Animal Chromosome Analysis (G-banded) Service
- I-FISH Service
- AAV Biodistribution Analysis (RNA ISH)
- Molecular Karyotyping (aCGH)
- Droplet Digital PCR (ddPCR) Service
- Digital ISH Image Quantification and Statistical Analysis
- SCE (Sister Chromatid Exchange) Analysis
- Biosample Services
- Histology Services
- Exosome Research Services
- In Vitro DMPK Services
-
In Vivo DMPK Services
- Pharmacokinetic and Toxicokinetic
- PK/PD Biomarker Analysis
- Bioavailability and Bioequivalence
- Bioanalytical Package
- Metabolite Profiling and Identification
- In Vivo Toxicity Study
- Mass Balance, Excretion and Expired Air Collection
- Administration Routes and Biofluid Sampling
- Quantitative Tissue Distribution
- Target Tissue Exposure
- In Vivo Blood-Brain-Barrier Assay
- Drug Toxicity Services
CYP and UGT Reaction Phenotyping Assay
Creative Bioarray helps provide CYP and UGT reaction phenotyping assay for CYP1A2, CYP2B6, CYP2C8, CYP3A4, UGT1A1, UGT2B7, or other isoforms on clients' requests. By identifying the enzymes responsible for metabolism, it points out the direction for identifying potential drug-drug interactions.
Reaction Phenotyping Assay
- Why Reaction Phenotyping Assay?
- Response phenotyping is the process of identifying enzymes responsible for drug metabolism. It allows to understand the role of metabolic enzymes in drug clearance by quantifying the contribution of major enzymes to the total drug clearance rate.
- Subtle changes in the enzyme activity will alter the rate of clearance and lead to negative results, that is, higher or lower plasma concentrations, which may result in unexpected toxicity or loss of efficacy. If multiple drugs interact with a single CYP or UGT form, then the possibility of a potentially harmful drug-drug interaction is increased.
- Reaction phenotyping studies identify which enzymes are catalyzing the main elimination pathways.
Brief Protocol
Reaction phenotyping assays, which are increasingly important in drug discovery efforts, are designed to identify specific enzymes involved in the metabolism of drug candidates. The basis of CYP and UGT reaction phenotyping is to understand enzyme kinetics of a specific enzyme metabolic pathway and determine its contribution to total metabolic clearance. A variety of assays can be utilized including those using recombinant cDNA-expressed human CYPs and UGTs.
- The test compound is incubated with the expressed single CYP450 enzyme or uridine diphosphate glucuronyl transferase at 37°C.
- For the cytochrome P450 response, the cofactor NADPH is used to initiate the response. The solvent terminates the reaction.
- After centrifugation, the supernatant was analyzed by LC-MS/MS. Monitor the disappearance of the test compound over 45 minutes.
- The test compound was also incubated in the presence of a cDNA expression control preparation (without enzyme). In the presence of the expressed enzyme, the compound disappearance curve is corrected to any disappearance observed in the presence of the control formulation. This ensures that only the metabolism of the allogeneic body is determined.
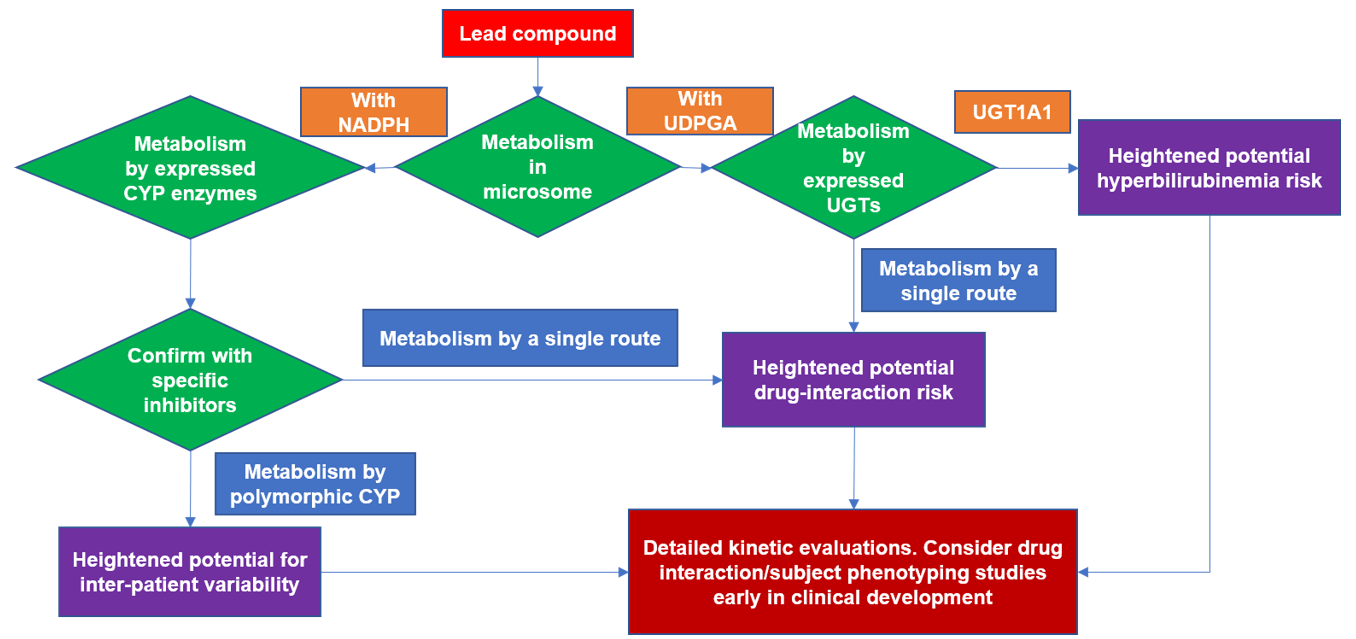 Figure 1. Flowchart for typical reaction phenotyping evaluation of a lead drug candidate.
Figure 1. Flowchart for typical reaction phenotyping evaluation of a lead drug candidate.
| CYP and UGT reaction phenotyping assay | |
| Test Concentration | 5μm (or other concentrations) |
| Enzymes | CYP1A2, CYP2B6, CYP2C8, CYP3A4, UGT1A1, UGT2B7 (or other isoforms) |
| Time Points | 0, 5, 15, 30, 45, 60, 120 minutes |
| Number of Replicates | n=3 per time point |
| Negative Controls | cDNA expressed control preparation (no CYP450 or UGT enzyme present) |
| Analysis Method | LC-MS/MS |
Quotation and Ordering
If you have any special needs or questions regarding our services, please feel free to contact us. We look forward to cooperating with you in the future.
Reference
- Chen Y, Fretland AJ, et al. Utility of intersystem extrapolation factors in early reaction phenotyping and the quantitative extrapolation of human liver microsomal intrinsic clearance using recombinant cytochromes P450. Drug Metab Dispos. 2011 Mar; 39 (3):373-82.
Explore Other Options
For research use only. Not for any other purpose.

