- You are here: Home
- Services
- ISH/FISH Services
- Transgene Mapping (Locus Amplification & Sequencing)
Services
-
Cell Services
- Cell Line Authentication
- Cell Surface Marker Validation Service
-
Cell Line Testing and Assays
- Toxicology Assay
- Drug-Resistant Cell Models
- Cell Viability Assays
- Cell Proliferation Assays
- Cell Migration Assays
- Soft Agar Colony Formation Assay Service
- SRB Assay
- Cell Apoptosis Assays
- Cell Cycle Assays
- Cell Angiogenesis Assays
- DNA/RNA Extraction
- Custom Cell & Tissue Lysate Service
- Cellular Phosphorylation Assays
- Stability Testing
- Sterility Testing
- Endotoxin Detection and Removal
- Phagocytosis Assays
- Cell-Based Screening and Profiling Services
- 3D-Based Services
- Custom Cell Services
- Cell-based LNP Evaluation
-
Stem Cell Research
- iPSC Generation
- iPSC Characterization
-
iPSC Differentiation
- Neural Stem Cells Differentiation Service from iPSC
- Astrocyte Differentiation Service from iPSC
- Retinal Pigment Epithelium (RPE) Differentiation Service from iPSC
- Cardiomyocyte Differentiation Service from iPSC
- T Cell, NK Cell Differentiation Service from iPSC
- Hepatocyte Differentiation Service from iPSC
- Beta Cell Differentiation Service from iPSC
- Brain Organoid Differentiation Service from iPSC
- Cardiac Organoid Differentiation Service from iPSC
- Kidney Organoid Differentiation Service from iPSC
- GABAnergic Neuron Differentiation Service from iPSC
- Undifferentiated iPSC Detection
- iPSC Gene Editing
- iPSC Expanding Service
- MSC Services
- Stem Cell Assay Development and Screening
- Cell Immortalization
-
ISH/FISH Services
- In Situ Hybridization (ISH) & RNAscope Service
- Fluorescent In Situ Hybridization
- FISH Probe Design, Synthesis and Testing Service
-
FISH Applications
- Multicolor FISH (M-FISH) Analysis
- Chromosome Analysis of ES and iPS Cells
- RNA FISH in Plant Service
- Mouse Model and PDX Analysis (FISH)
- Cell Transplantation Analysis (FISH)
- In Situ Detection of CAR-T Cells & Oncolytic Viruses
- CAR-T/CAR-NK Target Assessment Service (ISH)
- ImmunoFISH Analysis (FISH+IHC)
- Splice Variant Analysis (FISH)
- Telomere Length Analysis (Q-FISH)
- Telomere Length Analysis (qPCR assay)
- FISH Analysis of Microorganisms
- Neoplasms FISH Analysis
- CARD-FISH for Environmental Microorganisms (FISH)
- FISH Quality Control Services
- QuantiGene Plex Assay
- Circulating Tumor Cell (CTC) FISH
- mtRNA Analysis (FISH)
- In Situ Detection of Chemokines/Cytokines
- In Situ Detection of Virus
- Transgene Mapping (FISH)
- Transgene Mapping (Locus Amplification & Sequencing)
- Stable Cell Line Genetic Stability Testing
- Genetic Stability Testing (Locus Amplification & Sequencing + ddPCR)
- Clonality Analysis Service (FISH)
- Karyotyping (G-banded) Service
- Animal Chromosome Analysis (G-banded) Service
- I-FISH Service
- AAV Biodistribution Analysis (RNA ISH)
- Molecular Karyotyping (aCGH)
- Droplet Digital PCR (ddPCR) Service
- Digital ISH Image Quantification and Statistical Analysis
- SCE (Sister Chromatid Exchange) Analysis
- Biosample Services
- Histology Services
- Exosome Research Services
- In Vitro DMPK Services
-
In Vivo DMPK Services
- Pharmacokinetic and Toxicokinetic
- PK/PD Biomarker Analysis
- Bioavailability and Bioequivalence
- Bioanalytical Package
- Metabolite Profiling and Identification
- In Vivo Toxicity Study
- Mass Balance, Excretion and Expired Air Collection
- Administration Routes and Biofluid Sampling
- Quantitative Tissue Distribution
- Target Tissue Exposure
- In Vivo Blood-Brain-Barrier Assay
- Drug Toxicity Services
Transgene Mapping (Locus Amplification & Sequencing)
For transgenic cell lines and transgenic animals generated via many methods, the integration site is random and, in most cases, not known. Integration of a transgene can disrupt an endogenous gene, potentially interfering with interpretation of the phenotype. In addition, knowledge of where the transgene is integrated is important for planning of crosses between animals carrying a conditional allele and a given Cre allele in case the alleles are on the same chromosome. When the transgene insertion site is unknown, zygosity is determined by expensive quantitative PCR-based approach. These limitations often force investigators to maintain transgenic models or cell lines in a hemizygous state, which may lead to less than desired expression levels of the transgene and make it less efficient and more costly. Creative Bioarray has developed the Transgene Mapping (Locus Amplification & Sequencing) analysis, which is a powerful tool to sequence transgenes and their integration sites.
Transgene Mapping (Locus Amplification & Sequencing) analysis combines the next generation sequencing to selectively amplify and sequence the transgene and surrounding genomic region. This method not only uncovers insertion sites and sequences of integrated transgenes, but also enables detection of single nucleotide variations, structural variations, and transgene-transgene fusions.
Transgene Mapping (Locus Amplification & Sequencing) in the Cell Line Development Process
Transgene locus amplification & Sequencing presents a cost-effective and high-quality alternative to conventional approaches to control the quality of transgenic cells.
Transgene Mapping (Locus Amplification & Sequencing) analysis can:
- Identify the transgene integration site(s)
- Detect structural changes in the host DNA at the transgene integration site(s)
- Sequence the entire transgene and detect any sequence variants as well as structural changes within the transgene
- Provide an estimation of the transgene copy number
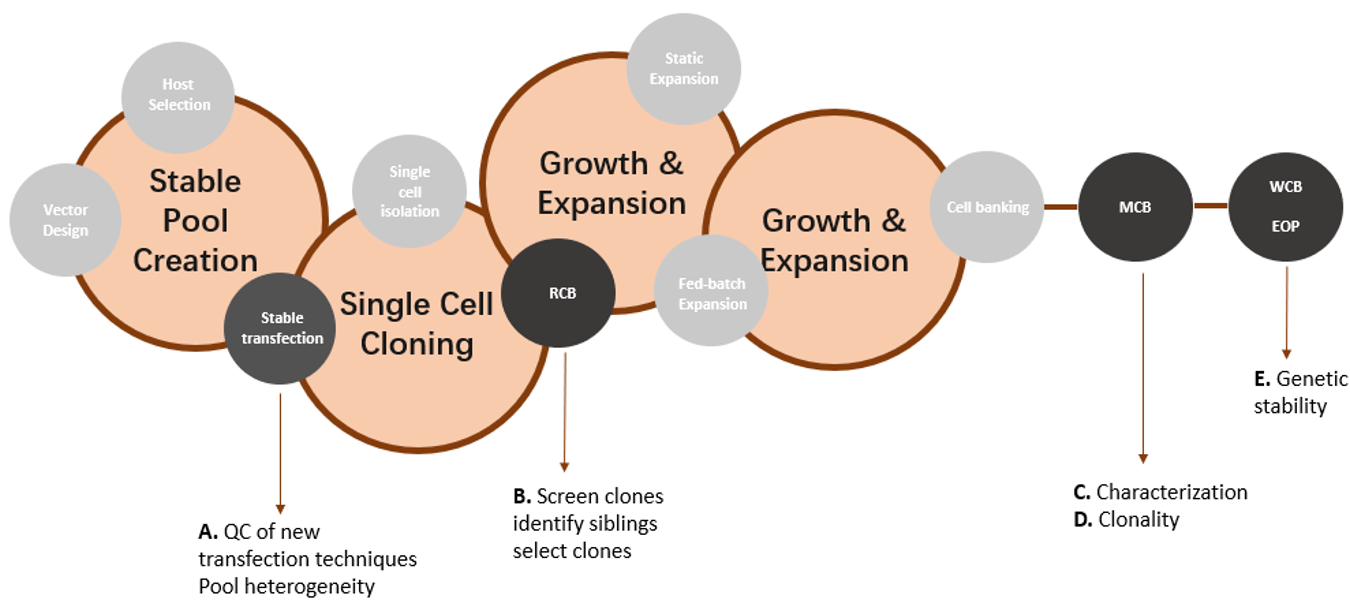 Figure 1: Overview of different stages of cell line development (in circles) and the purpose of Transgene Mapping (Locus Amplification & Sequencing) analyses (A-E).
Figure 1: Overview of different stages of cell line development (in circles) and the purpose of Transgene Mapping (Locus Amplification & Sequencing) analyses (A-E).
Transgene Mapping (Locus Amplification & Sequencing) in the Transgenic Animal Process:
Transgene Mapping (Locus Amplification & Sequencing) analysis can:
- Better correlation of phenotypes with transgene expression
- Ability to determine zygosity by genotyping assay and more cost-effective management of transgenic strains
- Enhanced predictability of transgene segregation when breeding and intercrossing
- Awareness of any potential disruption of the regulatory or coding region of a critical endogenous gene
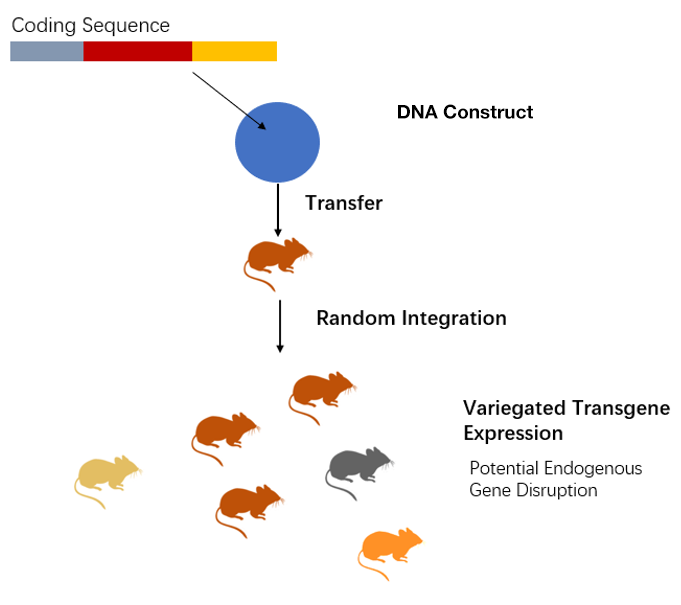 Figure 2. Transgene integration is random.
Figure 2. Transgene integration is random.
Features of Creative Bioarray's Transgene Mapping (Locus Amplification & Sequencing) Service:
- Identify the integration site(s)
- Identify the integrity of the transgene (sequence variants and structural variants)
- Identify the integrity of the host genome in the vicinity of the integration sites
- Identify the breakpoint sequences between the genome and the vector
- Identify vector-vector fusion reads to indicate the presence of concatemers
- Estimate the transgene copy number
- Allow the comprehensive genetic characterization of CHO cell lines
Advantages of Transgene Mapping (Locus Amplification & Sequencing) to conventional methods of transgene mapping

Deliverable
- Full scope support service-from sample preparation through to the final report
- Detailed report of insertion and transgene position
- Expert interpretation specific to a new transgenic line
Quotation and ordering
Our customer service representatives are available 24hr a day! We thank you for choosing Creative Bioarray at your preferred Transgene Mapping (Locus Amplification & Sequencing).
Explore Other Options
For research use only. Not for any other purpose.

