- You are here: Home
- Services
- Exosome Research Services
- Exosome Analysis
- Exosome Lipidomics Service
Services
-
Cell Services
- Cell Line Authentication
- Cell Surface Marker Validation Service
-
Cell Line Testing and Assays
- Toxicology Assay
- Drug-Resistant Cell Models
- Cell Viability Assays
- Cell Proliferation Assays
- Cell Migration Assays
- Soft Agar Colony Formation Assay Service
- SRB Assay
- Cell Apoptosis Assays
- Cell Cycle Assays
- Cell Angiogenesis Assays
- DNA/RNA Extraction
- Custom Cell & Tissue Lysate Service
- Cellular Phosphorylation Assays
- Stability Testing
- Sterility Testing
- Endotoxin Detection and Removal
- Phagocytosis Assays
- Cell-Based Screening and Profiling Services
- 3D-Based Services
- Custom Cell Services
- Cell-based LNP Evaluation
-
Stem Cell Research
- iPSC Generation
- iPSC Characterization
-
iPSC Differentiation
- Neural Stem Cells Differentiation Service from iPSC
- Astrocyte Differentiation Service from iPSC
- Retinal Pigment Epithelium (RPE) Differentiation Service from iPSC
- Cardiomyocyte Differentiation Service from iPSC
- T Cell, NK Cell Differentiation Service from iPSC
- Hepatocyte Differentiation Service from iPSC
- Beta Cell Differentiation Service from iPSC
- Brain Organoid Differentiation Service from iPSC
- Cardiac Organoid Differentiation Service from iPSC
- Kidney Organoid Differentiation Service from iPSC
- GABAnergic Neuron Differentiation Service from iPSC
- Undifferentiated iPSC Detection
- iPSC Gene Editing
- iPSC Expanding Service
- MSC Services
- Stem Cell Assay Development and Screening
- Cell Immortalization
-
ISH/FISH Services
- In Situ Hybridization (ISH) & RNAscope Service
- Fluorescent In Situ Hybridization
- FISH Probe Design, Synthesis and Testing Service
-
FISH Applications
- Multicolor FISH (M-FISH) Analysis
- Chromosome Analysis of ES and iPS Cells
- RNA FISH in Plant Service
- Mouse Model and PDX Analysis (FISH)
- Cell Transplantation Analysis (FISH)
- In Situ Detection of CAR-T Cells & Oncolytic Viruses
- CAR-T/CAR-NK Target Assessment Service (ISH)
- ImmunoFISH Analysis (FISH+IHC)
- Splice Variant Analysis (FISH)
- Telomere Length Analysis (Q-FISH)
- Telomere Length Analysis (qPCR assay)
- FISH Analysis of Microorganisms
- Neoplasms FISH Analysis
- CARD-FISH for Environmental Microorganisms (FISH)
- FISH Quality Control Services
- QuantiGene Plex Assay
- Circulating Tumor Cell (CTC) FISH
- mtRNA Analysis (FISH)
- In Situ Detection of Chemokines/Cytokines
- In Situ Detection of Virus
- Transgene Mapping (FISH)
- Transgene Mapping (Locus Amplification & Sequencing)
- Stable Cell Line Genetic Stability Testing
- Genetic Stability Testing (Locus Amplification & Sequencing + ddPCR)
- Clonality Analysis Service (FISH)
- Karyotyping (G-banded) Service
- Animal Chromosome Analysis (G-banded) Service
- I-FISH Service
- AAV Biodistribution Analysis (RNA ISH)
- Molecular Karyotyping (aCGH)
- Droplet Digital PCR (ddPCR) Service
- Digital ISH Image Quantification and Statistical Analysis
- SCE (Sister Chromatid Exchange) Analysis
- Biosample Services
- Histology Services
- Exosome Research Services
- In Vitro DMPK Services
-
In Vivo DMPK Services
- Pharmacokinetic and Toxicokinetic
- PK/PD Biomarker Analysis
- Bioavailability and Bioequivalence
- Bioanalytical Package
- Metabolite Profiling and Identification
- In Vivo Toxicity Study
- Mass Balance, Excretion and Expired Air Collection
- Administration Routes and Biofluid Sampling
- Quantitative Tissue Distribution
- Target Tissue Exposure
- In Vivo Blood-Brain-Barrier Assay
- Drug Toxicity Services
Exosome Lipidomics Service
Lipids are an essential composition of exosomal membranes. Exosomes contain several unique lipids compared with recipient cells, but these lipid compositions remain largely unknown. Exosome lipidomics analysis may help you identify components in exosomal lipids during disease progression, which may facilitate novel biomarker discovery for diseases diagnosis and prognosis.
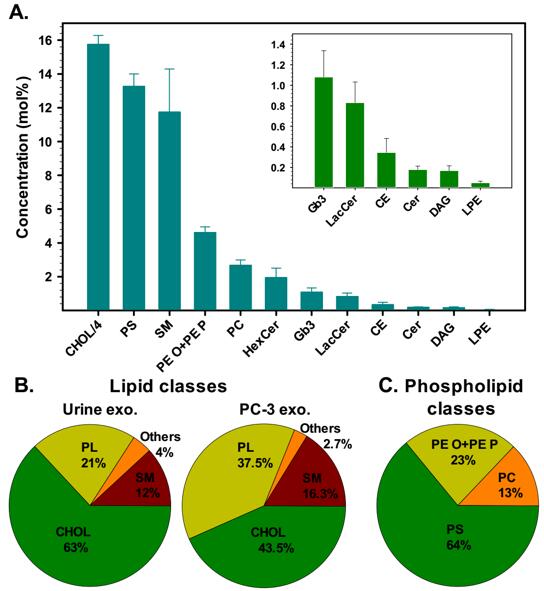 Figure 1 Lipid classes in urinary exosomes.
Figure 1 Lipid classes in urinary exosomes.
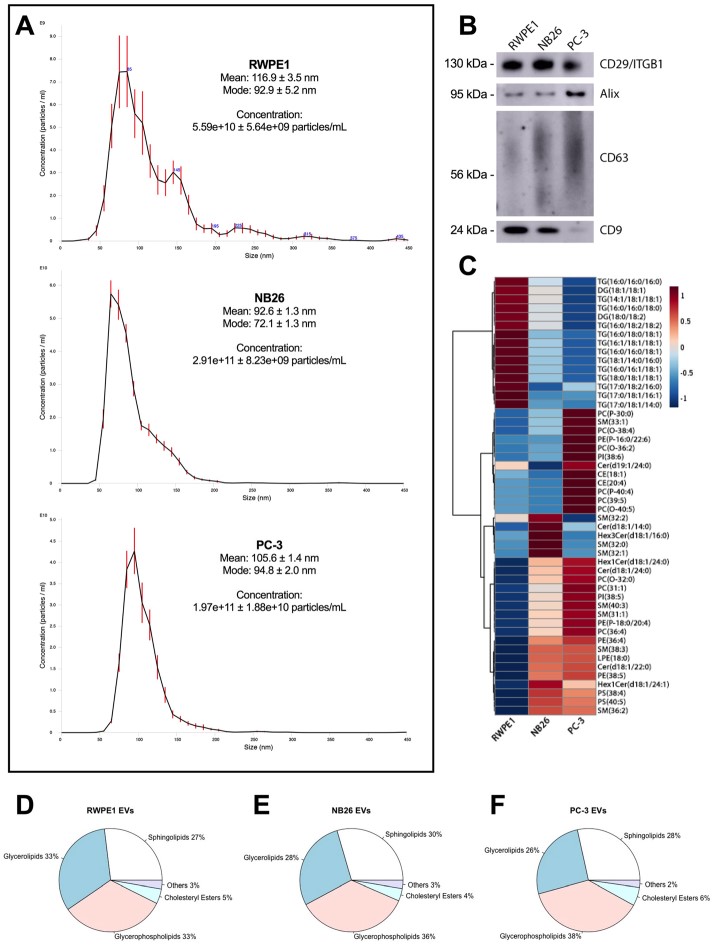 Figure 2 Extracellular vesicles (EVs) from tumourigenic cell lines have differences in molecular lipid composition compared to EVs from non-tumourigenic cells
Figure 2 Extracellular vesicles (EVs) from tumourigenic cell lines have differences in molecular lipid composition compared to EVs from non-tumourigenic cells
Current research has revealed lipid compositions of urinary exosomes in prostate cancer patients and healthy controls by using a high-throughput mass spectrometry method for quantitative lipidomics analysis. This study is the first to show the potential use of exosomal lipid species in urine as prostate cancer biomarkers.
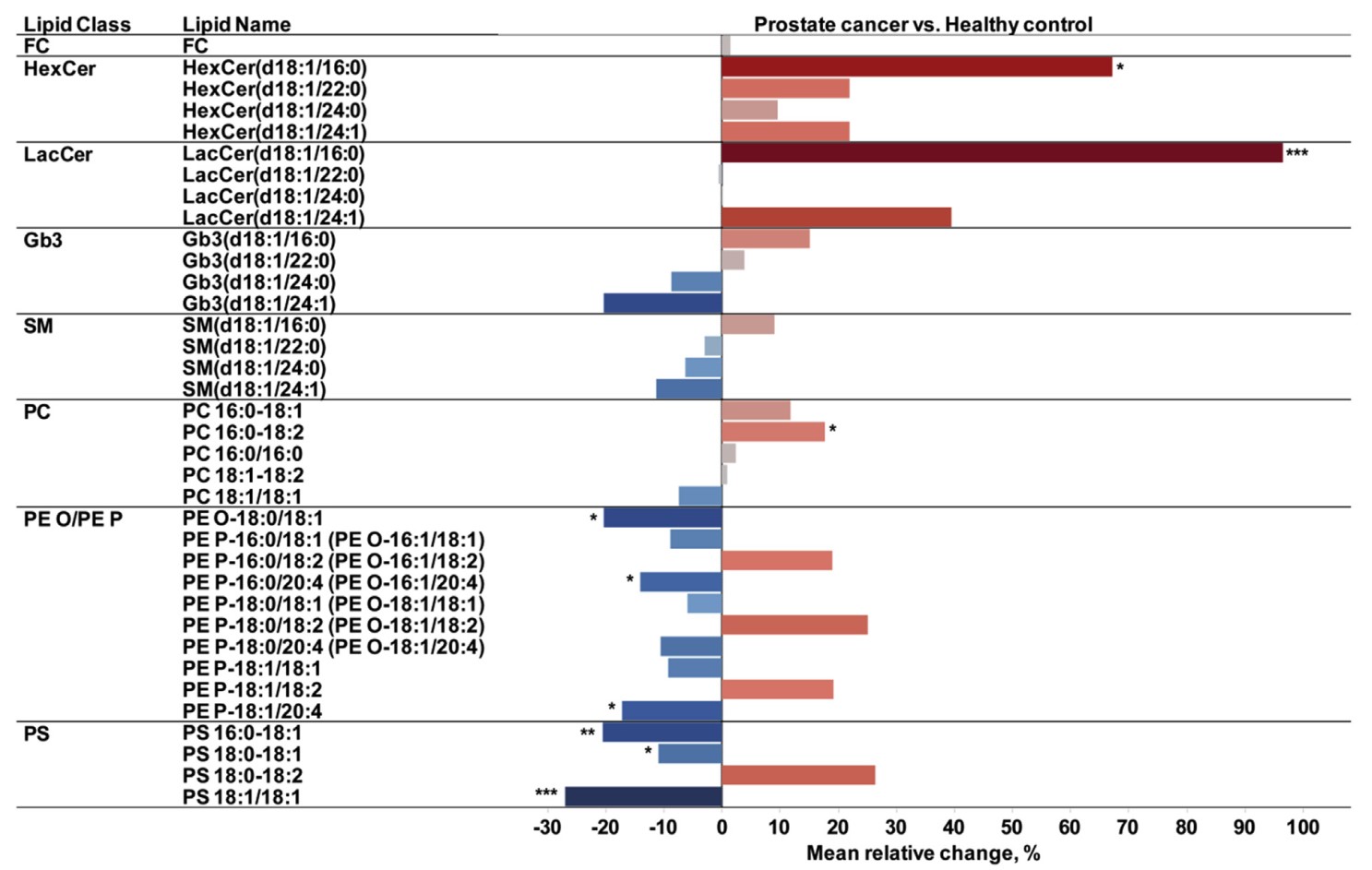 Figure 3 Comparison of molecular lipids in urinary exosomes from prostate cancer patients and healthy controls. Percent difference for each molecular lipid. Red and blue colors indicate that the increase or decrease of a molecular lipid, respectively, in the prostate cancer versus the control group. The relative difference is encoded by color intensity.
Figure 3 Comparison of molecular lipids in urinary exosomes from prostate cancer patients and healthy controls. Percent difference for each molecular lipid. Red and blue colors indicate that the increase or decrease of a molecular lipid, respectively, in the prostate cancer versus the control group. The relative difference is encoded by color intensity.
Creative Bioarray makes it easy to perform your exosome lipidomics research. Our high-throughput lipidomics analysis can provide lipid identification and quantification of your exosomes in detail. To request additional information or quotation for our exosome lipidomics service, please contact us. We look forward to working with you in the future.
References
- Skotland, T.; et al. Molecular lipid species in urinary exosomes as potential prostate cancer biomarkers. Eur J Cancer. 2017, 70: 122-132.
- Brzozowski, JS.; et al. Lipidomic profiling of extracellular vesicles derived from prostate and prostate cancer cell lines. Lipids Health Dis. 2018, 17(1): 211.
Explore Other Options
For research use only. Not for any other purpose.

