Featured Products
Our Promise to You
Guaranteed product quality, expert customer support

Retinoblastoma Cells
- Product List
- Background
- Applications
- Scientific Data
Retinoblastoma cells originate from the retinal cells, specifically from the photoreceptor cells or the retinal pigment epithelium. The development of retinoblastoma is typically associated with mutations in the RB1 gene, which encodes a protein called retinoblastoma protein (pRB). This protein acts as a tumor suppressor by helping to regulate the cell cycle and preventing excessive cell growth.
Cellular Biology of Retinoblastoma
- Characteristics of retinoblastoma cells. These cells exhibit heterogeneity in their morphology and genetic makeup, often resulting in varied behavior and response to treatments. Typically, retinoblastoma cells show high proliferation rates and possess a unique capability to evade apoptosis, contributing to their aggressive nature.
- Mechanisms of tumorigenesis. The tumorigenic process in retinoblastoma involves multiple pathways, with the inactivation of the RB1 gene being central. The loss of RB1 function allows for unchecked progression through the G1 phase of the cell cycle, facilitating excessive cell division. Furthermore, retinoblastoma cells may recruit various pro-survival signaling pathways, such as the PI3K/Akt and MAPK pathways, enhancing their resistance to chemotherapeutic agents. At the chromosomal level, retinoblastoma cells frequently display chromosomal abnormalities, including deletions and amplifications, particularly involving chromosomes 13 and 6.
Cancer Research
Retinoblastoma cells are used as a model system to study the fundamental processes of cancer development, including cell proliferation, apoptosis, and cell cycle control. The RB1 gene and its protein product, pRB, are central to understanding how cells regulate growth and prevent cancer.
Tumor Suppressor Mechanisms
The study of retinoblastoma cells helps in elucidating the function of tumor suppressor genes and their role in preventing tumorigenesis. This research can lead to the development of new therapeutic strategies that target pathways affected by tumor suppressor gene mutations.
Drug Discovery and Development
Retinoblastoma cells are valuable in testing new anticancer drugs and therapies. Because they represent a relatively simple and accessible model of childhood cancer, they can be used to screen for compounds that may inhibit tumor growth or induce cell death in cancer cells.
HRF1 Depletion Sensitizes Retinoblastoma Cells to Chemotherapeutic Drugs
UHRF1 (ubiquitin-like with PHD and ring finger domains 1) is highly expressed in various human cancers including retinoblastoma, and is associated with tumor-promoting effects such as inhibition of apoptosis and high proliferation. In an attempt to investigate how UHRF1 contributes to retinoblastoma development, the possibility that high UHRF1 expression was tested in retinoblastoma cells may endow the cells with the resistance against chemotherapeutic drugs used for retinoblastoma treatment in clinical settings. Etoposide (topoisomerase II inhibitor), camptothecin (topoisomerase I inhibitor), and carboplatin were used for dose-response and time-course studies on control and UHRF1-knockdown Y79 retinoblastoma cells (Fig. 1). UHRF1-depleted retinoblastoma cells showed a clear increase in sensitivity to the two topoisomerase inhibitors, whereas only modest increase in sensitivity to carboplatin was observed at high doses or longer treatment.
To examine if the increased drug sensitivity upon UHRF1 depletion accompanies higher apoptotic responses, Y79 knockdown cells were treated with etoposide and analyzed for several apoptosis markers. Sub-G1 and annexin V+ PI− early apoptotic populations were higher in UHRF1-depleted cells than in control cells (Fig. 2a, b). Upon etoposide treatment, both control and UHRF1-knockdown cells showed p53 induction and an increase in phosphorylation of upstream kinases in DNA damage responses. However, UHRF1-depleted cells displayed more robust apoptotic responses demonstrated by elevated levels of cleaved caspase-3 and poly(ADP-ribose) polymerase (PARP) (Fig. 2c, d). Interestingly, higher phosphorylation of histone H2AX at Ser139 (γH2AX) was detected in UHRF1-depleted cells, suggestive of higher susceptibility to DSB inducers such as etoposide. The higher sensitivity to etoposide was not due to the changes in topoisomerase expression in UHRF1-depleted cells. Similar apoptotic responses upon UHRF1 knockdown were observed for another retinoblastoma cell line, Weri-Rb1 (Fig. 2e). Consistent with the results shown in Fig. 1b, e, UHRF1-depleted Y79 cells also exhibited higher apoptosis in response to camptothecin (Fig. 2f).
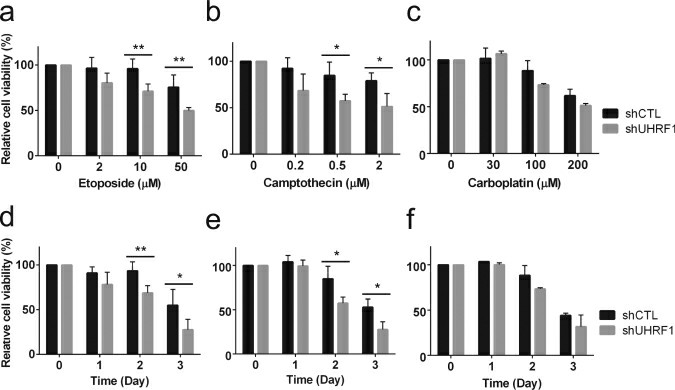 Fig. 1 (a-c) Dose-response study showing the relative sensitivity to drugs. Stable control knockdown (shCTL) and UHRF1-knockdown (shUHRF1) Y79 cells were exposed to various concentrations of chemotherapeutic drugs for 48 h, as indicated. (d-f) Time-course study showing the relative sensitivity to drugs. (He H, et al, 2018)
Fig. 1 (a-c) Dose-response study showing the relative sensitivity to drugs. Stable control knockdown (shCTL) and UHRF1-knockdown (shUHRF1) Y79 cells were exposed to various concentrations of chemotherapeutic drugs for 48 h, as indicated. (d-f) Time-course study showing the relative sensitivity to drugs. (He H, et al, 2018)
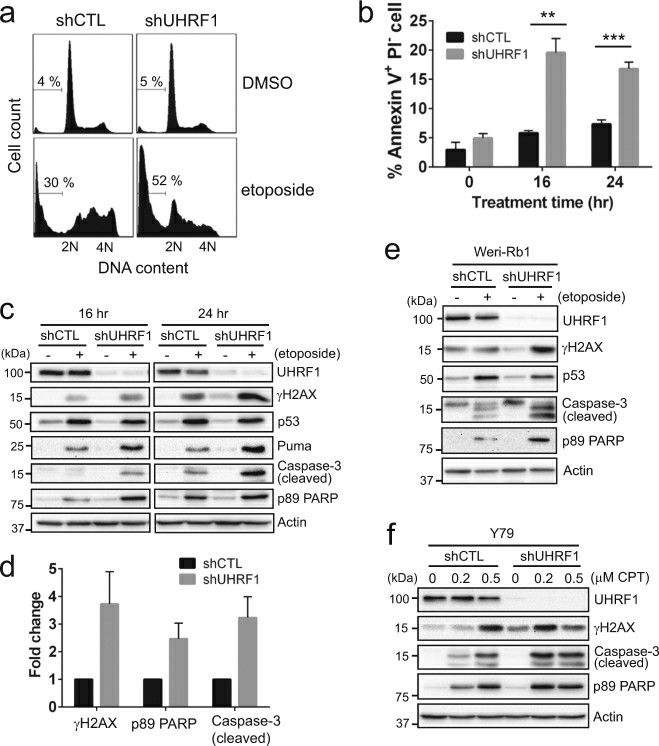 Fig. 2 Enhanced drug sensitivity upon UHRF1 depletion involves apoptotic cell death. (He H, et al, 2018)
Fig. 2 Enhanced drug sensitivity upon UHRF1 depletion involves apoptotic cell death. (He H, et al, 2018)
High GSDME Expression Switched Apoptosis to Pyroptosis in Retinoblastoma Cells
Pyroptosis is mediated by the gasdermin (GSDM) family of GSDMA, GSDMB, GSDMC, DFNA5/GSDME, and DFNB59. GSDME is a member of the GSDM family and can be cleaved by activated caspase-3 to form an active N-terminal domain (GSDME-N), which is inserted into the cell membrane to form pores, causing cell swelling and lysis, the release of inflammatory substances, and pyroptosis.
GSDME has recently been considered a tumor suppressor that promotes tumor cell death by inducing pyroptosis. Carboplatin was used to induce the death of Y79 and WERI-RB-1 cells, and the IC50 in Y79 cells was significantly higher than that in WERI-RB-1 cells (Fig. 3a and Table 1). First, observation of the death morphology of the three cell lines showed that the main form of cell death was apoptosis in Y79 cells but pyroptosis in ARPE-19 cells, and some apoptosis occurred in WERI-RB-1 cells (Fig. 3b). In the case of carboplatin-induced cell death in Y79, WERI-RB-1, and ARPE-19 cells, a decrease in Bcl-2, an increase in Bax, and significant increases in activated caspase-9, activated caspase-7, and caspase-3 were observed. GSDME-N was present in cells expressing the GSDME protein (Fig. 3c-i). Flow cytometry was performed to detect cell death. After treatment with carboplatin for 24 h, the ratio of FITC mono staining to FITC and PI double-positive staining was higher in Y79 cells than in WERI-RB-1 cells, and the ratio of FITC mono staining to FITC and PI double-positive staining was higher in WERI-RB-1 cells than in ARPE-19 cells (Fig. 3j and k). A large amount of LDH was detected in the cell supernatant of WERI-RB-1 and ARPE-19 cells (Fig. 3l). However, in this study, cleavage-activated caspase-9 was also present in the WB of the normal group, but no activated caspase-7 or caspase-3 was observed downstream—a finding that requires further study.
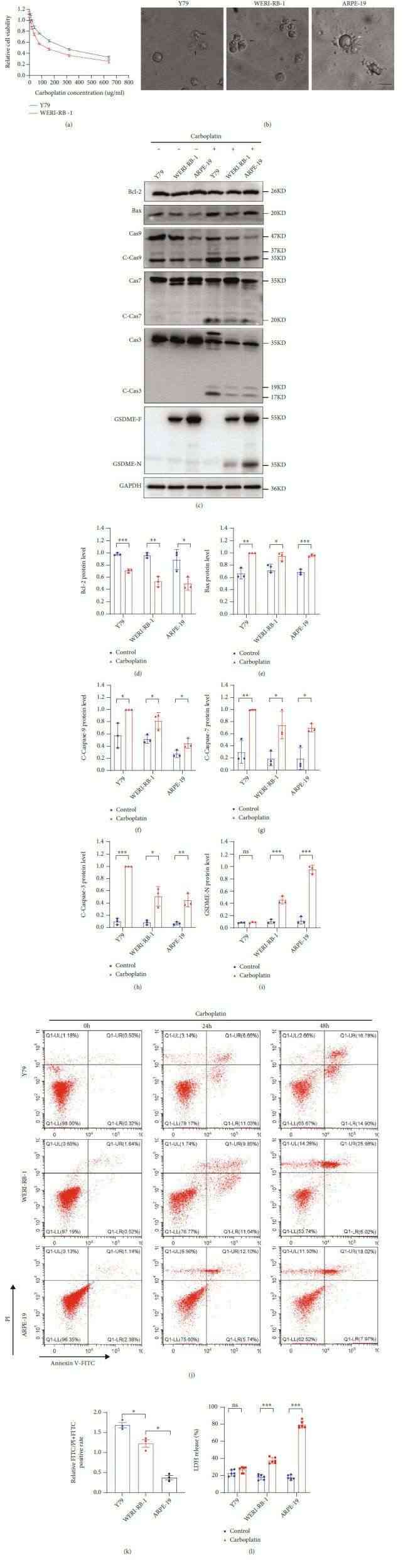 Fig. 3 (a) CCK-8 assay of cell viability. (b) The death morphologies of the three cell lines were observed using an Olympus microscope. (c–i) GSDME-N and pathway proteins were tested by WB in three different cell lines. (j and k) The FITC and PI staining rates of the three cell lines were determined by flow cytometry and quantitative maps were drawn after treatment with carboplatin for 24 h. (l) LDH levels in the cell supernatant were detected by ELISA. (Li F, et al., 2022)
Fig. 3 (a) CCK-8 assay of cell viability. (b) The death morphologies of the three cell lines were observed using an Olympus microscope. (c–i) GSDME-N and pathway proteins were tested by WB in three different cell lines. (j and k) The FITC and PI staining rates of the three cell lines were determined by flow cytometry and quantitative maps were drawn after treatment with carboplatin for 24 h. (l) LDH levels in the cell supernatant were detected by ELISA. (Li F, et al., 2022)
Table 1. IC50 values and statistical analyses of carboplatin treatments in RB cell lines. (Li F, et al., 2022)
| IC50 Carboplatin | |
| Y79 | 121.9 (99.04 to 151.3) |
| WERI-RB-1 | 65.53 (45.89 to 77.18) |
Description: Human cell line derived from retinoblastoma.
Description: The line exhibited a hypodiploid karyotype initially, and an epithelial morphology.
Description: Human retinoblastoma cell line derived from a female Caucasian. The cells are semi-adherent...
Description: Human retinoblastoma cell line derived from a male patient. Bilateral retinoblastoma...

