- You are here: Home
- Services
- ISH/FISH Services
- FISH Applications
- mtRNA Analysis (FISH)
Services
-
Cell Services
- Cell Line Authentication
- Cell Surface Marker Validation Service
-
Cell Line Testing and Assays
- Toxicology Assay
- Drug-Resistant Cell Models
- Cell Viability Assays
- Cell Proliferation Assays
- Cell Migration Assays
- Soft Agar Colony Formation Assay Service
- SRB Assay
- Cell Apoptosis Assays
- Cell Cycle Assays
- Cell Angiogenesis Assays
- DNA/RNA Extraction
- Custom Cell & Tissue Lysate Service
- Cellular Phosphorylation Assays
- Stability Testing
- Sterility Testing
- Endotoxin Detection and Removal
- Phagocytosis Assays
- Cell-Based Screening and Profiling Services
- 3D-Based Services
- Custom Cell Services
- Cell-based LNP Evaluation
-
Stem Cell Research
- iPSC Generation
- iPSC Characterization
-
iPSC Differentiation
- Neural Stem Cells Differentiation Service from iPSC
- Astrocyte Differentiation Service from iPSC
- Retinal Pigment Epithelium (RPE) Differentiation Service from iPSC
- Cardiomyocyte Differentiation Service from iPSC
- T Cell, NK Cell Differentiation Service from iPSC
- Hepatocyte Differentiation Service from iPSC
- Beta Cell Differentiation Service from iPSC
- Brain Organoid Differentiation Service from iPSC
- Cardiac Organoid Differentiation Service from iPSC
- Kidney Organoid Differentiation Service from iPSC
- GABAnergic Neuron Differentiation Service from iPSC
- Undifferentiated iPSC Detection
- iPSC Gene Editing
- iPSC Expanding Service
- MSC Services
- Stem Cell Assay Development and Screening
- Cell Immortalization
-
ISH/FISH Services
- In Situ Hybridization (ISH) & RNAscope Service
- Fluorescent In Situ Hybridization
- FISH Probe Design, Synthesis and Testing Service
-
FISH Applications
- Multicolor FISH (M-FISH) Analysis
- Chromosome Analysis of ES and iPS Cells
- RNA FISH in Plant Service
- Mouse Model and PDX Analysis (FISH)
- Cell Transplantation Analysis (FISH)
- In Situ Detection of CAR-T Cells & Oncolytic Viruses
- CAR-T/CAR-NK Target Assessment Service (ISH)
- ImmunoFISH Analysis (FISH+IHC)
- Splice Variant Analysis (FISH)
- Telomere Length Analysis (Q-FISH)
- Telomere Length Analysis (qPCR assay)
- FISH Analysis of Microorganisms
- Neoplasms FISH Analysis
- CARD-FISH for Environmental Microorganisms (FISH)
- FISH Quality Control Services
- QuantiGene Plex Assay
- Circulating Tumor Cell (CTC) FISH
- mtRNA Analysis (FISH)
- In Situ Detection of Chemokines/Cytokines
- In Situ Detection of Virus
- Transgene Mapping (FISH)
- Transgene Mapping (Locus Amplification & Sequencing)
- Stable Cell Line Genetic Stability Testing
- Genetic Stability Testing (Locus Amplification & Sequencing + ddPCR)
- Clonality Analysis Service (FISH)
- Karyotyping (G-banded) Service
- Animal Chromosome Analysis (G-banded) Service
- I-FISH Service
- AAV Biodistribution Analysis (RNA ISH)
- Molecular Karyotyping (aCGH)
- Droplet Digital PCR (ddPCR) Service
- Digital ISH Image Quantification and Statistical Analysis
- SCE (Sister Chromatid Exchange) Analysis
- Biosample Services
- Histology Services
- Exosome Research Services
- In Vitro DMPK Services
-
In Vivo DMPK Services
- Pharmacokinetic and Toxicokinetic
- PK/PD Biomarker Analysis
- Bioavailability and Bioequivalence
- Bioanalytical Package
- Metabolite Profiling and Identification
- In Vivo Toxicity Study
- Mass Balance, Excretion and Expired Air Collection
- Administration Routes and Biofluid Sampling
- Quantitative Tissue Distribution
- Target Tissue Exposure
- In Vivo Blood-Brain-Barrier Assay
- Drug Toxicity Services
mtRNA Analysis (FISH)
Mammalian mitochondria contain a small circular genome, which encodes 2 ribosomal RNAs, 22 tRNAs, and 13 essential protein subunit. The transcription and translation machineries in the mitochondrial matrix require both mitochondrion-encoded RNAs and nucleus encoded protein and RNA factors. Mitochondrial RNAs (mtRNAs) are first transcribed as polycistronic transcripts from both H-strand and L-strand, and the transcripts are dissected by RNA processing enzyme into individual rRNAs, tRNAs, and mRNAs. These rRNAs and tRNAs together with translational factors imported from cytosol then synthesize the proteins from the mRNAs.
Mitochondrial RNA homeostasis is one of the key elements in regulating mitochondrial functions. Mitochondria have to quickly respond to external stimuli and are constantly going through fusion and fission, which is directly linked to their biosynthesis and requires fast changes of their gene expression. Much is understood about mtRNA synthesis and processing. But distinct localization of mitochondrial RNA (mtRNA) and the molecular mechanism are poorly understood.
It is well known among researchers that mitochondrial genetic or primary mitochondrial disorders contribute to mitochondrial dysfunction. Beyond the evaluation of mtDNA, levels of mitochondrial RNA (mtRNA) and mitochondrial regualtory genes can provide additional information regarding the risk of clinically improtant mitochondrial damage. Fluorescence in situ hybridization (FISH) provides a reliable tool for charaterizing the fate and location of mtRNA. Creative Bioarray offers the detection of mtRNA in your specific studies.
 Fig 1. Mammalian mitochondria
Fig 1. Mammalian mitochondria
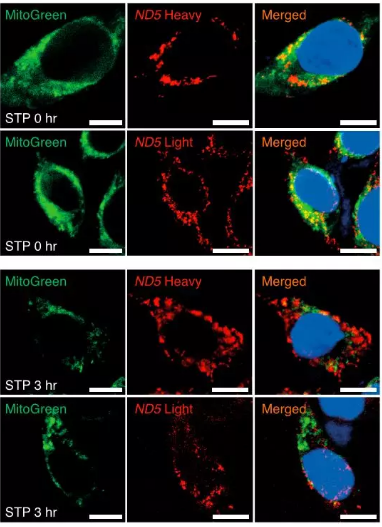 Fig 2. Cytosolic release of mtRNAs can be visualized in situ using RNA ISH. Bar indicates 10 µm.
Fig 2. Cytosolic release of mtRNAs can be visualized in situ using RNA ISH. Bar indicates 10 µm.
Kim Y, Park J, Kim S, et al. PKR senses nuclear and mitochondrial signals by interacting with endogenous double-stranded RNAs[J]. Molecular cell, 2018, 71(6): 1051-1063. e6.
Application:
- Mitochodrial Function Study
- dsRNA Related Study
Features:
- Accurate - In Situ Detection Service - Custom design your probe
- Value - We focus on the quality of our service and all supported by competitive pricing
- Efficiency - We are able to provide the fastest turnaround time of any supplier in the industry
Creative Bioarray offers custom mtRNA Analysis services for your scientific research as follows:
- Probe design
- Probe synthesis
- Sample (Cells/FFPE/Others) preparation
- Imaging
- Data analysis
References
- Kim Y, et al. PKR senses nuclear and mitochondrial signals by interacting with endogenous double-stranded RNAs[J]. Molecular cell, 2018, 71(6): 1051-1063. e6.
- Birkus G.; et al. Assessment of mitochondrial toxicity in human cells treated with tenofovir: comparison with other nucleoside reverse transcriptase inhibitors[J]. Antimicrobial agents and chemotherapy, 2002, 46(3): 716-723.
- Bobrowicz A J.; et al. Polyadenylation and degradation of mRNA in mammalian mitochondria: a missing link?[J]. 2008.
- Nicolson G L. Mitochondrial dysfunction and chronic disease: treatment with natural supplements[J]. Integrative Medicine: A Clinician's Journal, 2014, 13(4): 35.
- Liu P.; et al. Mammalian mitochondrial RNAs are degraded in the mitochondrial intermembrane space by RNASET2[J]. Protein & cell, 2017, 8(10): 735-749.
Explore Other Options
For research use only. Not for any other purpose.

