Featured Products
Our Promise to You
Guaranteed product quality, expert customer support

ONLINE INQUIRY
COS-1
Cat.No.: CSC-C3432
Species: Monkey
Source: kidney
Morphology: fibroblast-like, adherent monolayers
Culture Properties: monolayer
- Specification
- Background
- Scientific Data
- Q & A
- Customer Review
Viruses: ELISA: reverse transcriptase negative; PCR: EBV -, HCV -, HBV -, HIV -, HTLV-I/II -, SMRV -, SMRV -
COS-1 cell line, derived from fibroblasts from the kidney of African green monkeys. These cells are tightly attached to the culture surface under standard culture conditions and grow in a spreading monolayer. The COS-1 cell line was obtained by transfection with SV40 viruses containing mutations encoding the wild-type T antigen but having defective mutations at the start site. They have integrated a single copy of an intact early region of the SV40 genome and are capable of expressing the T antigen, making them an ideal choice for transfections that require the SV40 T-antigen expression vector. Ideal for transfection with vectors requiring SV40 T antigen expression. This feature not only simplifies the construction of recombinant vectors, but also allows the COS-1 cell line to support replication of temperature-sensitive A209 viruses at 40°C, as well as replication of pure SV40 with a deletion of the early region. The COS-1 cell line does not have stringent limitations on the amount of DNA to be inserted or the genomic DNA sequences to be used, and allows for the rapid validation of cDNA-positive clones and the analysis of sequence mutations, providing a powerful tool for gene function studies. Mutations, providing a powerful tool for gene function studies. Therefore, the COS-1 cell line is not only suitable as a packaging host for the production of high-quality recombinant lentiviral vectors, but also requires no additional purification steps to produce high-titer viral vectors.
Functional Analysis of Abnormal Antithrombin Protein (N87D)
Antithrombin (AT) is a plasma serine protease inhibitor that inactivates coagulation factors. Inherited AT deficiency is an autosomal dominant thrombotic disorder caused by various mutations in SERPINC1. A novel missense mutation was discovered by Kamijima et al. in a female patient with AT deficiency. Analysis of the patient's DNA revealed a missense mutation due to a single A to G base substitution at c.259 in exon 2 of SERPINC1. This base change is predicted to alter the amino acid at position 87 of the AT protein from asparagine (Asn, N) to aspartic acid (Asp, D) (N87D). To assess this mutation, wild-type and mutant AT cDNA were transfected into COS-1 cells and cultured for 48 hours. Post-culture, cells were lysed, separating supernatant and pellet, and AT protein identification was performed using an AT antigen. Results indicated the relative levels of the N87D mutant AT antigen detected in the cell culture and in the supernatant using ELISA were 67 ± 16 and 11 ± 5%, respectively. The antigen level in the cell culture was therefore slightly lower, but not significantly different. The antigen level in the supernatant was significantly lower (Fig. 1). Meanwhile, the distribution of AT in cells was detected by immunofluorescence. The results showed that both wild type and mutant type AT showed a network fluorescence distribution in the cytoplasm, and there was no significant difference in their fluorescence signal localization (Fig. 2). In conclusion, their data demonstrate that the N87D mutation in AT leads to AT deficiency, characterized by reduced levels of AT antigen and activity.
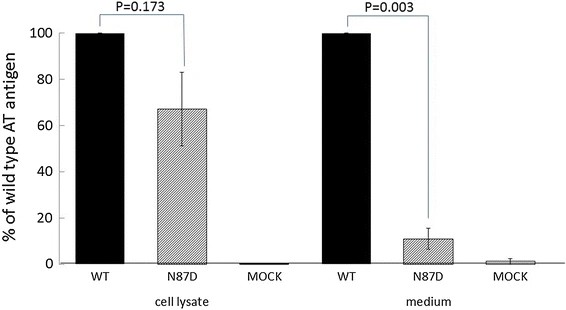 Fig. 1. The levels of recombinant AT antigen in cell lysates and culture medium of COS-1 cells transfected with either wild-type or N87D mutant AT expression vectors (Kamijima S, Sekiya A, et al., 2018).
Fig. 1. The levels of recombinant AT antigen in cell lysates and culture medium of COS-1 cells transfected with either wild-type or N87D mutant AT expression vectors (Kamijima S, Sekiya A, et al., 2018).
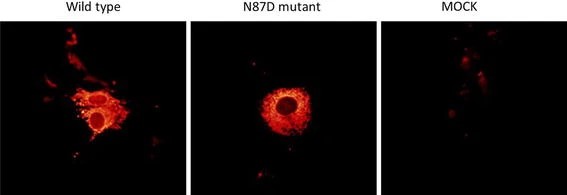 Fig. 2. Fluorescence immunostaining (Kamijima S, Sekiya A, et al., 2018).
Fig. 2. Fluorescence immunostaining (Kamijima S, Sekiya A, et al., 2018).
Novel Application of Macrolampis sp2 Firefly Luciferase for Intracellular pH-Biosensing in Mammalian Cells (GOS-1)
Firefly luciferase-based bioluminescent sensors are among the most popular sensors available. Firefly luciferase is sensitive to pH changes, exhibiting a significant red shift under acidic conditions. Gabriel's team localized luciferase from the Macrolampis sp2 firefly to either the cytoplasm or nucleus in mammalian cells (GOS-1) to observe pH changes during biological activity. Images were captured using dichroic mirrors that detected bioluminescence intensities below and above 590 nm, distinguishing between green and red emissions. At pH 8.0, green emission intensified in both the cytoplasm and nucleus, while at pH 6.5, red emission was predominant (Fig. 3), indicating that luciferase's bioluminescent response is similar to pH alterations. These findings are consistent with previous observations in E. coli expressing the Macrolampis sp2 luciferase. They further assessed the bioluminescence intensity of green and red emissions using the ratio (F5/F4) (Fig. 4), revealing that green light increases under more basic conditions. At acidic pH, the cytoplasm appeared slightly greener than the nucleus. These results suggest that this firefly luciferase can serve as a diagnostic tool for measuring intracellular pH variations.
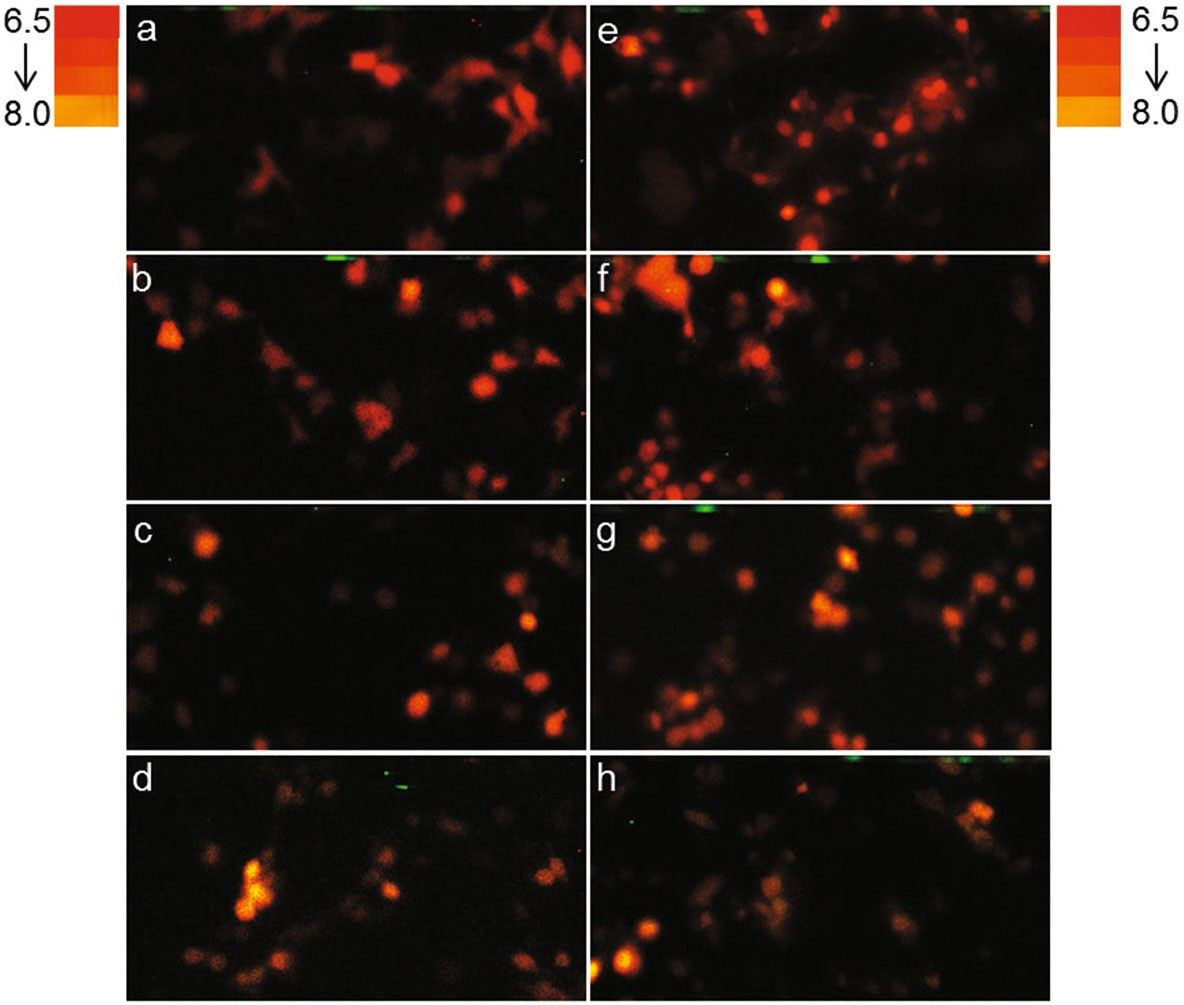 Fig. 3. Bioluminescence color in COS-1 cells under different pH conditions (Gabriel GVM, Yasuno R, et al., 2019).
Fig. 3. Bioluminescence color in COS-1 cells under different pH conditions (Gabriel GVM, Yasuno R, et al., 2019).
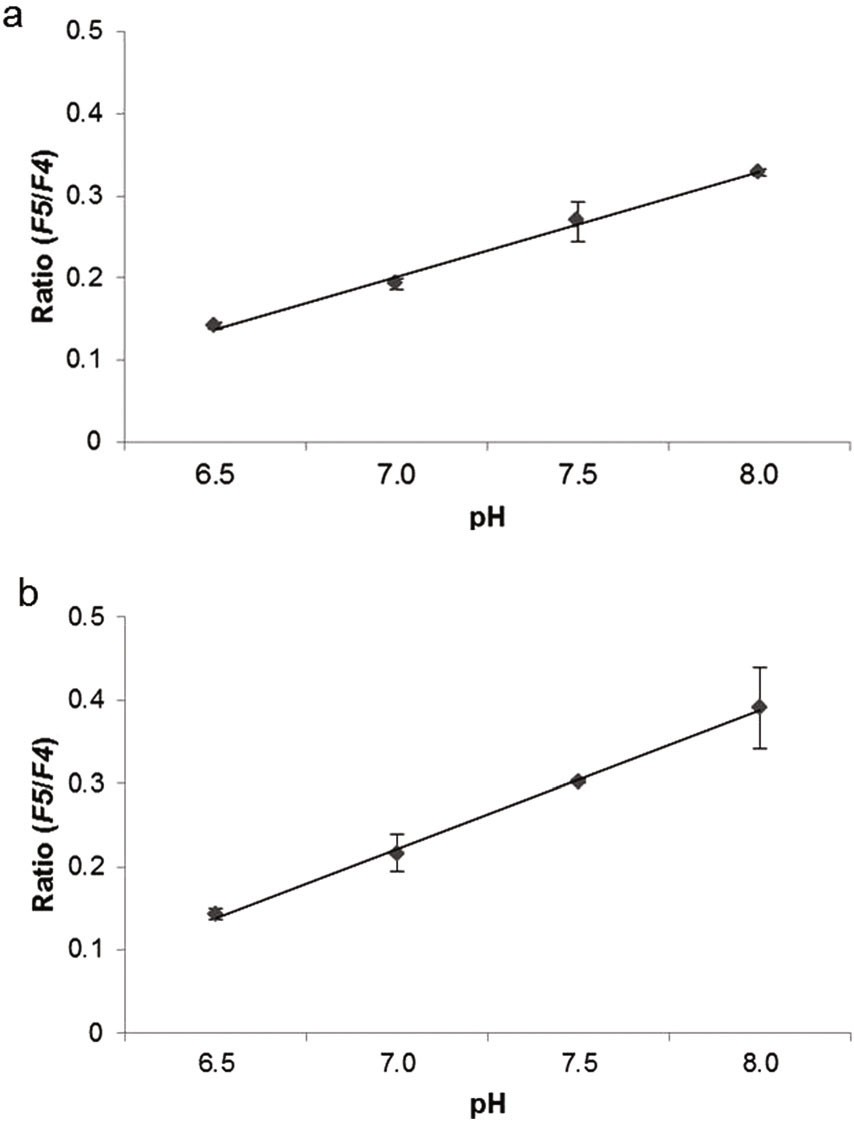 Fig. 4. Bioluminescence intensity variation of green and red (Gabriel GVM, Yasuno R, et al., 2019).
Fig. 4. Bioluminescence intensity variation of green and red (Gabriel GVM, Yasuno R, et al., 2019).
Ask a Question
Write your own review
- You May Also Need