BS-C-1
Cat.No.: CSC-C9342L
Species: Chlorocebus pygerythrus (Vervet monkey)
Source: Kidney
Morphology: epithelial
Culture Properties: adherent
- Specification
- Background
- Scientific Data
- Q & A
- Customer Review
Strain: Cercopithecus aethiops
Virus Susceptibility: SV40; poliovirus 1; vesicular stomatitis (Indiana)
Production: keratin
Histopathology: normal
Note: the cells are positive for keratin by immunoperoxidase staining
The BS-C-1 cell line, created by primary culture from kidney tissue from African green monkeys (Cercopithecus aethiops), was screened by Hopps in 1961. They are epithelial cells, polygonal and pressed tightly to one another like paving stones.
The BS-C-1 cell line is sensitive to many viruses such as Poliovirus type 1, SV40 virus, and Vesicular stomatitis virus, which allows them to be used in the testing of viruses. They are also ideal hosts for viral vector transfection, particularly SV40 vectors. In cell culture, the BS-C-1 cell line can be grown in MEM media with 10% foetal bovine serum and doubled after approximately 72 hours. In terms of gene expression, BS-C-1 cells were most likely to have expressed keratins, suggesting that they were epithelial cells.
The BS-C-1 cell line has applications in virology research, vaccine research, antiviral drug discovery, cell signalling research, and cell membrane transport studies. For toxicology research, the BS-C-1 cell line can also be used to assess chemical effects on cells, and measure toxicity and potential targets for chemicals through markers of cell survival, morphology and metabolism.
Nucleotide Substitutions in D10R are Associated with Enhanced Replication of Spontaneous Mutants in BS-C-1 Cells.
The modified vaccinia virus Ankara (MVA) is an attenuated virus used as a smallpox vaccine and in clinical trials for other vaccines. Known for its safety due to its inability to replicate in most mammalian cells. However, a spontaneous mutation in MVA enhanced its replication in monkey BS-C-1 cells. The impact of this mutation on the immunogenicity of MVA vectors remains unknown. Erez et al. isolated variants (MVA47.1) with enhanced replication in BS-C-1 cells through blind passaging of MVA and characterized the effect of spontaneous mutations in the D10 decapping enzyme on viral replication.
Initially, MVA was passaged in BS-C-1 cells to identify mutations extending the host range. A total of 10 rounds of low multiplicity infections were conducted, as shown in Fig. 1A. Large plaques began appearing post the second round and dominated after the sixth (Fig. 1B). Viruses from three plaques in the 10th round were cloned as BSC P1.1, BSC P1.2, BSC P1.3, BSC P2.1, BSC P2.2, and BSC P2.3. As a control, CEFs were inoculated with the starting MVA and viruses from three plaques were clonally purified to produce CEF P1.1, CEF P1.2, and CEF P1.3. At 48 hours, the yield of BS-C-1-adapted viruses was 2 logs higher than non-adapted MVAs (Fig. 1C) and exhibited similar replication ability to MVA 47.1 in BS-C-1 cells (Fig. 1D). Four cloned viruses were expanded in BS-C-1 cells, and their DNA was analyzed by sequencing. Each virus had a single nucleotide change in the D10R ORF, leading to either an alanine-to-threonine mutation at amino acid 226 or a histidine-to-tyrosine mutation at amino acid 233. Mutations at position 226 were found in clones from passage series 1, which also exhibited unique large deletions and one A14L ORF substitution. Mutations at position 233 were in clones from series 2 without other alterations. The MVA 47.1 virus had a different D10 mutation at amino acid 25. Comparison showed that D10 mutations alone were enough for enhanced replication in BS-C-1 cells.
 Fig. 1. Blind passage of MVA in BS-C-1 cells (Erez N, Wyatt LS, et al., 2021).
Fig. 1. Blind passage of MVA in BS-C-1 cells (Erez N, Wyatt LS, et al., 2021).
VACV Produces Higher Levels of Detectable dsRNA Than ECTV
Most orthopoxviruses, including vaccinia virus (VACV), contain genes in the E3L and K3L families. The protein products of these genes have been shown to combat PKR, a host defense pathway. Although VACV and ECTV exhibit high genomic similarity, their host ranges differ significantly. VACV is known for producing significant dsRNA, triggering antiviral responses. ECTV shows efficient PKR evasion despite its truncated K3L, possibly by producing less dsRNA. Frey's team aims to understand how ECTV manages to avoid the PKR pathway efficiently without a functional K3L gene and explores the role of dsRNA in immune evasion.
ECTV and VACV, while closely related, exhibit significant differences in host range and in vivo pathogenesis, resulting in varying levels of inflammation in mice when infected via the footpad route. Notably, ECTV lacks a functional K3 protein, unlike VACV. Since dsRNA is a crucial pathogen-associated risk signal to the mammalian immune system, it was thought that dsRNA production might differ between cells infected with ECTV and VACV. Armed with the monoclonal antibody J2, which specifically binds to dsRNA, fluorescence microscopy showed that most VACV-infected BS-C-1 cells carried dsRNA, but not many ECTV-infected cells. Replication-inhibiting drugs such as AraC were able to eradicate the detectable dsRNA from VACV-infected cells, implying that dsRNA is most likely formed by post-replication (Fig. 2A). Flow cytometry showed that VACV-infected BS-C-1 cells produced more dsRNA than ECTV-treated cells (Figs. 2B and 2C). An RNA dot blot with purified total RNA further validated more dsRNA in VACV-infected cells than in ECTV-infected ones (Fig. 2D). This result confirmed ECTV produced less dsRNA, consistent with previous findings. Notably, host beta-actin mRNA was absent in RNA from cells infected with either virus (Fig. 2D).
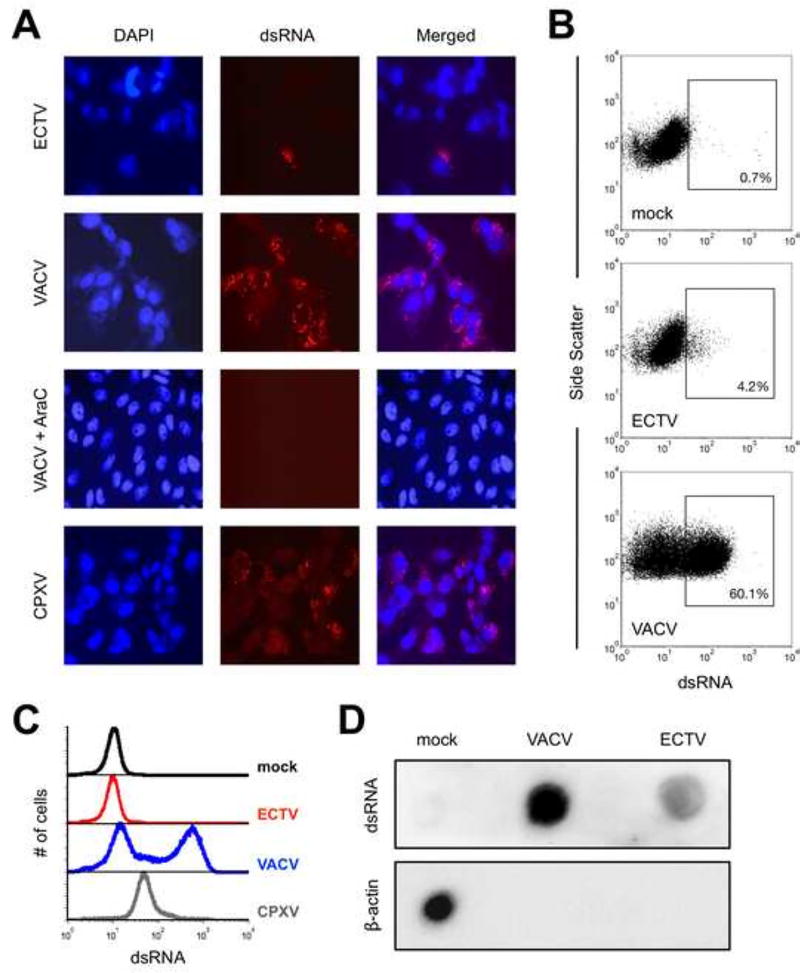 Fig. 2. The accumulation of dsRNA greatly varies after infection with different orthopoxviruses (Frey TR, Lehmann MH, et al., 2017).
Fig. 2. The accumulation of dsRNA greatly varies after infection with different orthopoxviruses (Frey TR, Lehmann MH, et al., 2017).
Ask a Question
Write your own review
- You May Also Need
- Adipose Tissue-Derived Stem Cells
- Human Neurons
- Mouse Probe
- Whole Chromosome Painting Probes
- Hepatic Cells
- Renal Cells
- In Vitro ADME Kits
- Tissue Microarray
- Tissue Blocks
- Tissue Sections
- FFPE Cell Pellet
- Probe
- Centromere Probes
- Telomere Probes
- Satellite Enumeration Probes
- Subtelomere Specific Probes
- Bacterial Probes
- ISH/FISH Probes
- Exosome Isolation Kit
- Human Adult Stem Cells
- Mouse Stem Cells
- iPSCs
- Mouse Embryonic Stem Cells
- iPSC Differentiation Kits
- Mesenchymal Stem Cells
- Immortalized Human Cells
- Immortalized Murine Cells
- Cell Immortalization Kit
- Adipose Cells
- Cardiac Cells
- Dermal Cells
- Epidermal Cells
- Peripheral Blood Mononuclear Cells
- Umbilical Cord Cells
- Monkey Primary Cells
- Mouse Primary Cells
- Breast Tumor Cells
- Colorectal Tumor Cells
- Esophageal Tumor Cells
- Lung Tumor Cells
- Leukemia/Lymphoma/Myeloma Cells
- Ovarian Tumor Cells
- Pancreatic Tumor Cells
- Mouse Tumor Cells