ONLINE INQUIRY

Canine Small Intestinal Epithelial Cells
Cat.No.: CSC-C9125J
Species: Dog
Source: Small Intestine; Intestine
Cell Type: Epithelial Cell
- Specification
- Background
- Scientific Data
- Q & A
- Customer Review
Canine small intestine epithelial cells are isolated from canine small intestinal tissue. The primary function of the small intestine in digestion includes absorbing food and distributing nutrients to the entire body. Canine small intestine epithelial cells develop their typical polygonal and irregular shapes when they establish a tightly packed single-layer monolayer in vitro. The intestinal mucosa contains these cells as primary components which block bacteria and viruses from entering the body while residing within the mucosal layer. Active and passive transport mechanisms allow these cells to become the primary site for the absorption of water and essential nutrients like electrolytes, amino acids and fatty acids. These cells release multiple enzymes and hormones such as insulin-like growth factor (IGF) and gastrin which help control digestive processes and metabolic functions. During immune responses these cells modulate the intestinal immune environment through their interactions with immune cells such as macrophages. Researchers utilize this cell line to study intestinal disorders such as inflammatory bowel disease alongside intestinal infections.
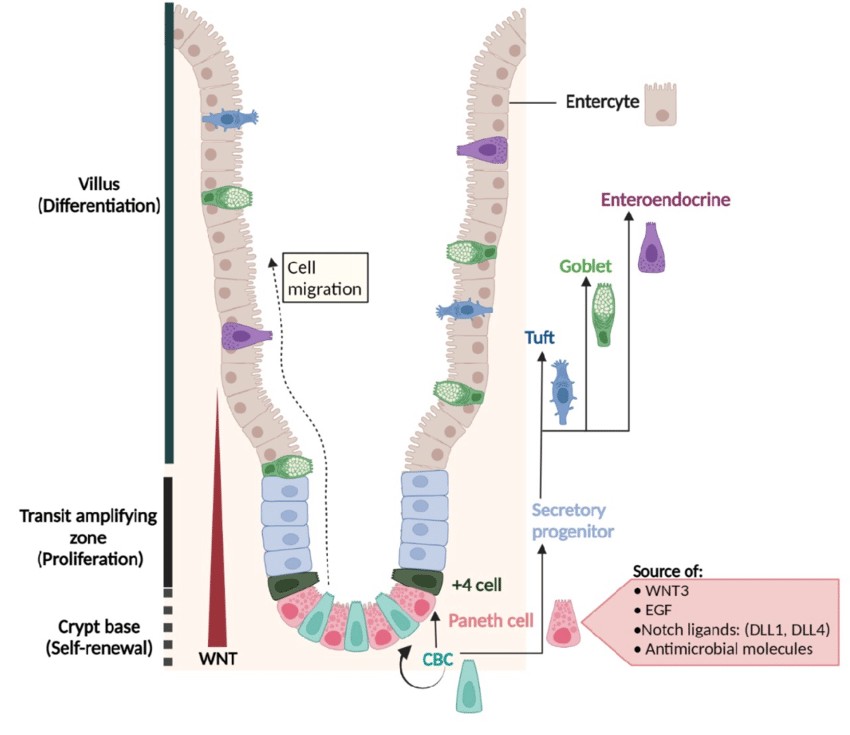 Fig. 1. Architecture of the small-intestinal epithelium and its regulatory signals (Brischetto C, 2022).
Fig. 1. Architecture of the small-intestinal epithelium and its regulatory signals (Brischetto C, 2022).
Analysis of Pathway Enrichment of the Response of Canine Intestinal Epithelial Cells Treated by Sulforaphane
Sulforaphane (SFN), a compound from cruciferous vegetables, is known for its anti-inflammatory and antioxidant properties, particularly beneficial for intestinal health. It can modulate inflammatory responses and enhance intestinal cell regeneration. Wang's team explored SFN's effects on canine small intestinal epithelial cells, focusing on its molecular impact through transcriptome analysis to understand its role in modulating biological processes involved in inflammation and oxidative stress.
To pinpoint SFN's regulatory transcriptional pathways, they conducted a transcriptome analysis on canine small intestinal epithelial cells, using SFN (4 μM) and Vehicle groups. They filtered the fold change of FPKM for valid data and performed enrichment analysis with GSEA 4.1.0. Key enrichments in the SFN group included the Inflammatory response, Lipid biosynthesis, Oxidative stress response, and T-cell immunity pathways (Fig. 1A), with the "Inflammatory response" pathway having the highest significance. Up-regulated genes focused on Protein digestion, PPAR signaling, and Fatty acid biosynthesis in KEGG (Fig. 1B and C). Down-regulated genes involved Cytokine interaction, MAPK signaling, and IL-17 pathways (Fig. 1D and E). Up-regulated DEGs also enriched in tissue and protein structure pathways, whereas down-regulated ones related to cytoarchitecture (Fig. 1F).
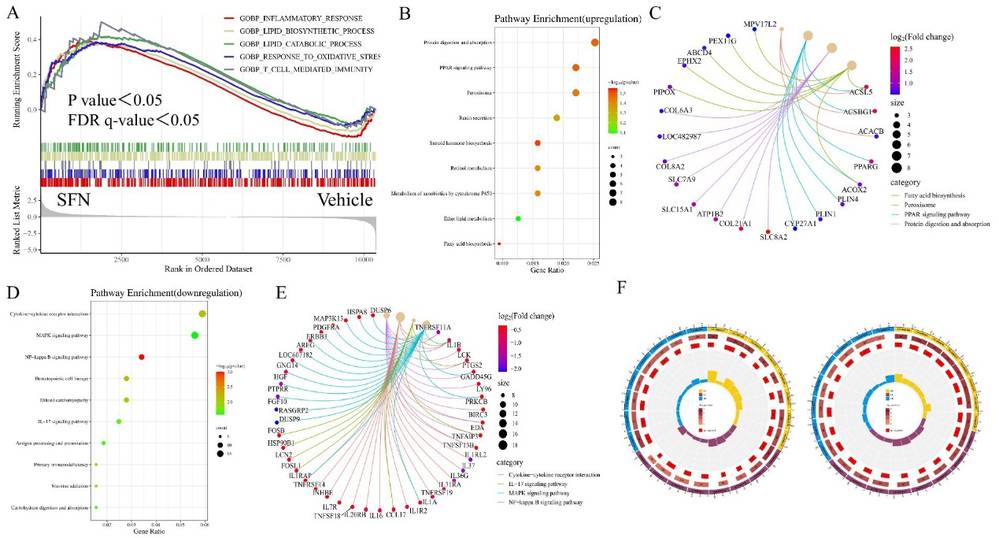 Fig. 1. GSEA, KEGG and GO enrichment analysis of DEGs. Showing GSEA enrichment plot, enrichment bubble plot, Cnetplot and GO enrichment circle plot (Li K, Yan J, et al., 2024).
Fig. 1. GSEA, KEGG and GO enrichment analysis of DEGs. Showing GSEA enrichment plot, enrichment bubble plot, Cnetplot and GO enrichment circle plot (Li K, Yan J, et al., 2024).
MLT and Cell Growth, Survival, and KEGG Enrichment in cIECs
Melatonin (MLT), a hormone produced by the pineal gland, plays a crucial role in regulating inflammation in mammals. Hong's team investigates how melatonin influences gene expression in canine ileum epithelial cells (Canine small intestinal epithelial cells, cIECs), focusing on the modulation of inflammatory genes ZAP70 and CD40 through histone modifications.
To evaluate the protective effects of a 10 μM MLT extract on cIECs, cell viability and apoptosis were assessed. No significant differences in cell numbers or caspase 3/7 activity, an apoptosis indicator, were observed between MLT-treated and control groups (Fig. 2A and B). Transcriptome analysis highlighted genes significantly affected by MLT (Fig. 2C and D), with GSEA identifying primary immunodeficiency and NF-kappa B signaling as key enriched pathways (Fig. 2E). No major changes were noted in cell cycle or proliferation genes (Fig. 2F). Functional annotation and pathway enrichment analysis (Fig. 3A-C) supported these findings, while the PPI network (Fig. 3D and E) showed the complex interactions and regulatory effects influenced by MLT on cIECs.
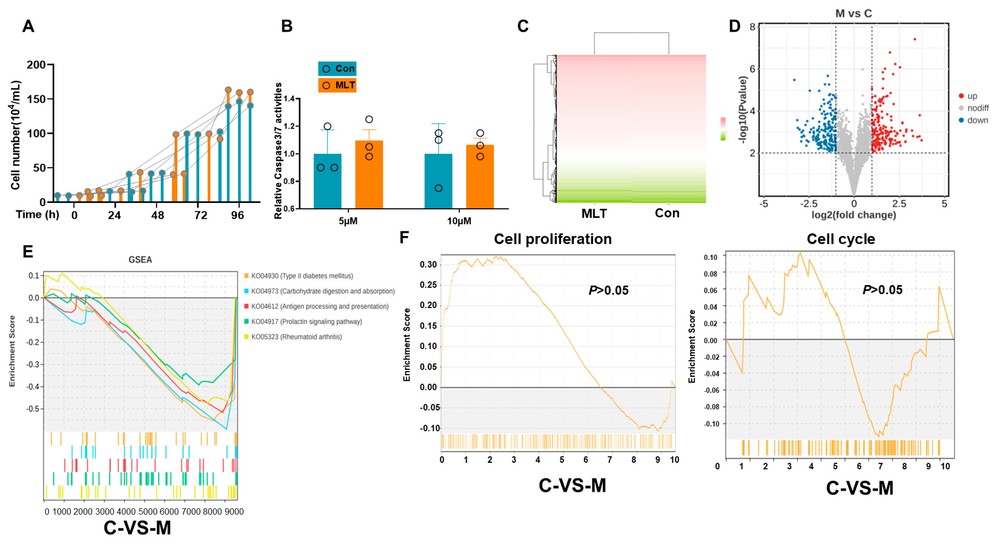 Figure 2. MLT treatment does not affect cell survival and growth in canine epithelial cells (Hong J, Adam SY, et al., 2025).
Figure 2. MLT treatment does not affect cell survival and growth in canine epithelial cells (Hong J, Adam SY, et al., 2025).
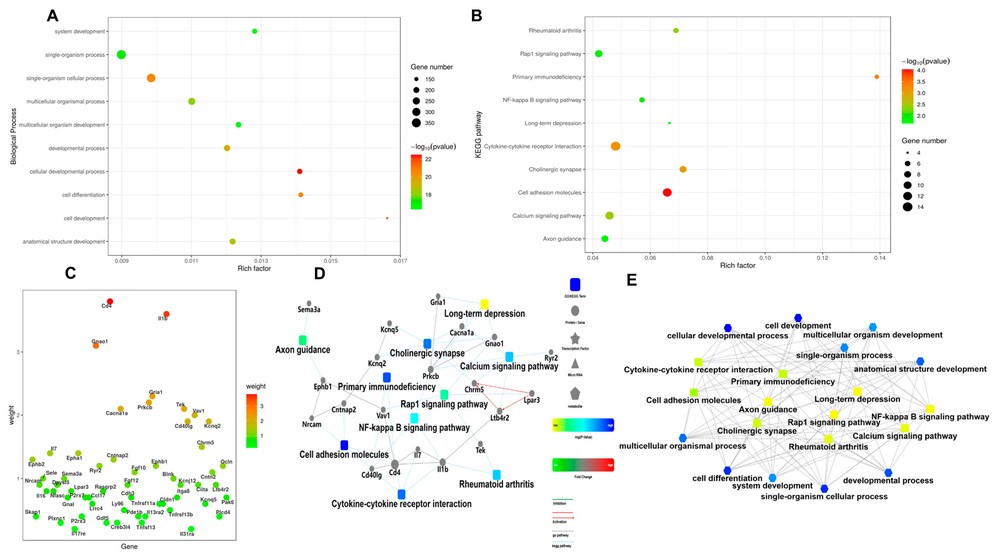 Fig. 3. Functional annotation pathway enrichment result (Hong J, Adam SY, et al., 2025).
Fig. 3. Functional annotation pathway enrichment result (Hong J, Adam SY, et al., 2025).
Ask a Question
Write your own review


