Overview of In Situ Hybridization
- What is ISH
- Main Advantages
- Probe Preparation
- Work Flow
- ISH Technology Optimization
- ISH Types
- ISH Applications
What is ISH
In Situ Hybridization (ISH)
ISH is a technology of detecting nucleotide sequences in cells, tissues, and tissue sections. It works by integrating a nucleotide probe that matches the sequence of target DNA or RNA with a stable hybrid for localization and detection. These probes are typically labelled with radioactive, fluorescent or other non-radioactive elements. Images are produced by a variety of developmental techniques, including radioautography, fluorescence microscopy, and immunohistochemistry. ISH reveals cell types without the destruction of tissue, and also reveals the relationship between the distribution of specific nucleic acids and their encoded products and cellular structure. Since its first publication in 1969, this technique has become a powerful instrument for research and clinical diagnosis, and is now routinely used in viral infection, gene expression, cytogenetics and prenatal diagnosis.
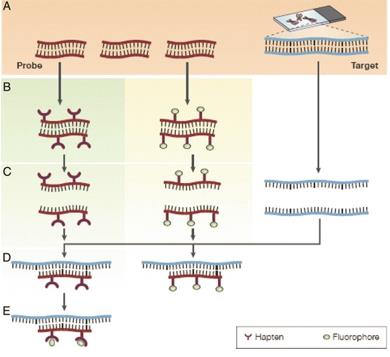
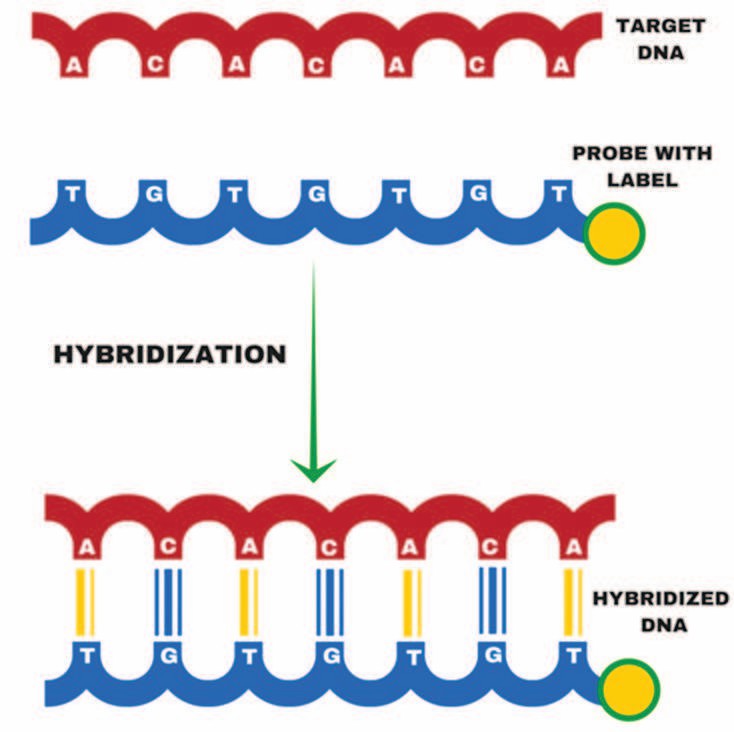 Fig. 1. Hybridization (Ehtisham, M., 2016)
Fig. 1. Hybridization (Ehtisham, M., 2016)
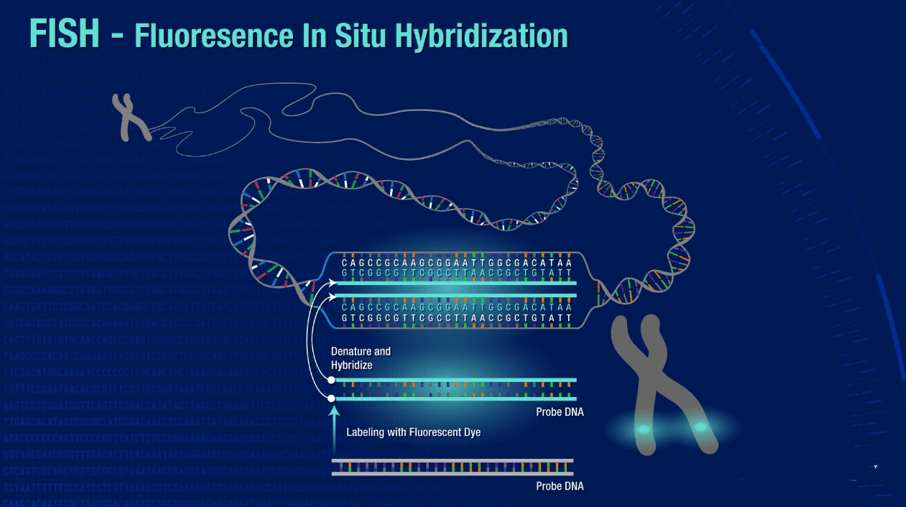
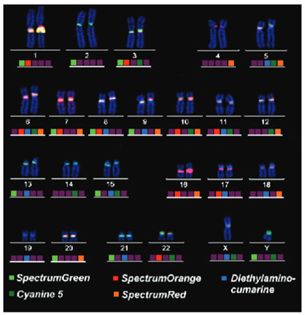
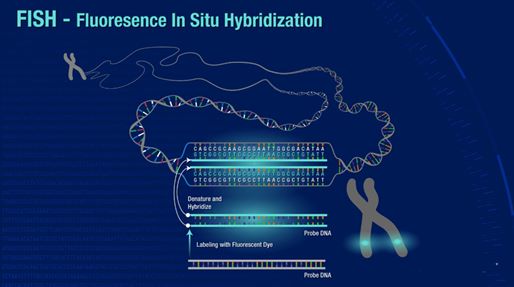
Fluorescence In Situ Hybridization (FISH)
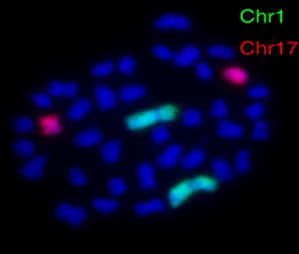
Among the in situ hybridization techniques, Fluorescence In Situ Hybridization (FISH) is an important branch. FISH is especially adept at molecular cytogenetic uses, including the diagnosis of chromosomal dysfunction and cellular biology. FISH is a technique that locates and detects a specific sequence of nucleic acids directly in the nuclei of intermediate chromosomes and interphase cells. If ISH was first uncovered using radioactive probes, FISH has set a new trend by using non-radioisotope probes that offer greater safety and resolution. FISH has served not only as a diagnostic tool for prenatal and systemic conditions, but has also been crucial in diagnosing and diagnosing hematological malignancies and solid tumors. In addition, FISH serves a complementary position in genomic studies such as chromosome microarrays (CMAs), and its application has even been extended to the use of super-resolution microscopy systems to visualize the organization of chromosomes in the nucleus.
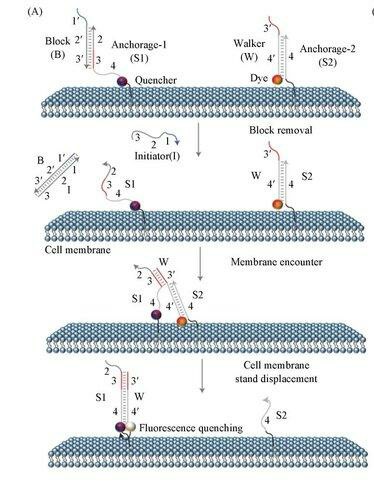
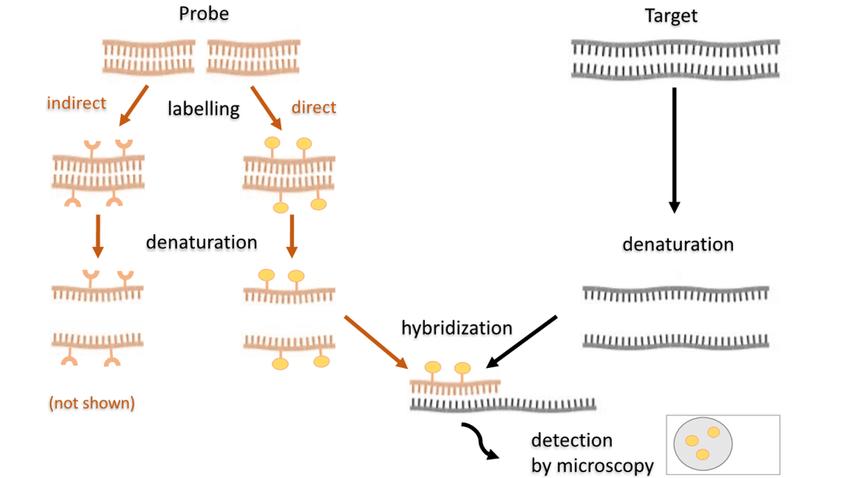 Fig. 2. Scheme of the fluorescence in situ hybridization using a fluorescently (directly) labeled probe (Kushkevych I, Hýžová B, et al., 2021).
Fig. 2. Scheme of the fluorescence in situ hybridization using a fluorescently (directly) labeled probe (Kushkevych I, Hýžová B, et al., 2021).
Main Advantages
ISH Technology:
- Strong localization specificity: ISH can detect specific DNA or RNA sequences directly in tissue sections or cells, pinpointing to the target gene or gene expression within the cell or tissue structure. And it can detect detect chromosomal abnormalities such as translocations, deletions and duplications.
- Flexibility: ISH technology allows for the selection of DNA or RNA probes that can be radioactively, fluorescently, or antigenically labeled with bases for colorimetric or fluorescent detection.
- Wide range of applications: It has been applied in many fields such as clinical cytogenetics, gene mapping, tumor biology and chromosome evolution studies.
FISH Technology:
- High sensitivity and specificity: FISH can detect target sequences with very low copy number and has high sequence specificity.
- Spatial resolution: FISH can localize target sequences at the single-cell level and visualize the distribution of molecules within the cell by fluorescence.
- Multiple detection: Multi-color FISH can detect multiple sequences simultaneously by displaying different colors within the same nucleus.
Probe Preparation
Probe types
| Type | Characteristics | Advantages | Disadvantages |
| double strand DNA probe | Include nick translation, random primer labeling, and PCR with labeled nucleotides, incorporating labeled nucleotides into DNA through various enzymatic actions. | Suitable for abundant targets. | Lower sensitivity compared to single-stranded probes, as double-stranded DNA tends to self-hybridize, reducing the concentration available for target hybridization. |
| single strand DNA probe | Covers a larger length (200-500 bp) and is prepared by reverse transcription PCR or chemical synthesis. | Simple PCR production with minimal starting material and flexible probe sequence selection, providing higher sensitivity and broader applications than double-stranded probes. | - |
| single strand RNA probe | Thermally stable and allows resistance to RNase degradation, generated in vitro during transcription. | Does not self-anneal in solution, avoids forming large molecular chains, and more easily penetrates cells or tissues, suitable for high-sensitivity detection. | - |
| oligonucleotide pro | Typically 20-50 bases long, produced by automated chemical synthesis. | Small size facilitates easy penetration into cells or tissues and RNase resistance; single-strand design prevents re-annealing. | Short length results in limited target region coverage. To enhance sensitivity, a mixture of probes complementary to different regions of the target molecule is often used. |
Probe labeling methods
Depending on the type of marker, it can be divided into radioisotope labeling and non-isotope labeling:
- Radioactive labeling includes the use of 32P, 33P, 35S, 3H, 125I and so on. This labeling method, although highly sensitive, has the disadvantages of long exposure time and low safety, and is rarely used at present.
- Non-isotopic labeling includes biotin, digoxin and fluorescein. These labeling methods are safe, stable and detectable by color development or fluorescence.
Probe detection
- Radioactive probe decetion: Radioisotope-labeled probes are used to visualize hybridization sites by radiographic autoradiography using X-ray film or liquid emulsion.
- Chemical detection: Compounds including biotin, fluorescein, digoxin, alkaline phosphatase, or bromodeoxyuridine are visualized by histochemistry or immunohistochemistry and observed by bright-field microscopy.
- Fluorescence decetion: Fluorescently labeled probes, visualized under a fluorescence microscope, are suitable for cytogenetic analysis, especially for the detection of chromosomal abnormalities.
Key considerations in probe preparation:
- Probe length: Excessive probe length can impede target accessibility and prolong hybridization time.
- Probe concentration: The typical concentration for DNA probes is 0.5-2 μg/ml. For unacetylated oligonucleotide probes, the concentration ranges from 50-200 ng/ml. The concentration can be adjusted upward for acetylated oligonucleotide probes.
- GC content and Tm value: The higher the GC content, and the higher the Tm value; in general, the hybridization effect of probes with high TM value is better than that of probes with low TM value.
- Complementary probe sequence: Self-complementary regions with more than three nucleotides within the probe should be avoided.
Work Flow
Probe preparation
Fixation: It is desirable to fix intermediate chromosome spreads with methanol/acetic acid. Cryostat sections can be fixed in 4% formaldehyde for about 30 minutes, or bouin solution, or by paraformaldehyde vapour fixation. In general, tissue remains are fixed in 10% formalin buffer, overnight processed by an automated tissue processor, and then embedded in paraffin.
Slicing: The embedded tissue samples were cut into thin slices. Tissue sections need to be dewaxed and hydrated by xylene and different concentrations of ethanol, and finally washed with PBS (phosphate buffer solution).
Digestion treatment: Pre-treating tissue with Proteinase K and others can enhance the ability of target nucleic acids to enter the tissue, improving hybridization efficiency.
Pre-hybridization: Incubate slices in hybridization buffer without probes to block sites that might non-specifically bind to probes, reducing background.
Hybridization: Hybridize the probe to the sample under appropriate conditions to bind the probe to the target sequence. If the probe is double-stranded, both the probe and the sample need to be denatured by heat and alkali treatment.
Post-hybridization washes: Remove unbound probes and non-specifically bound substances.
Detection: Select an appropriate detection method based on the type of probe label:
| Direct method | Indirect method |
| Reporter molecules 1. Enzyme, 2. Radioisotope or 3. Fluorescent markers are directly attached to the DNA or RNA | A hapten 1. Biotin, 2. Digoxigenin, or 3. Fluorescein are attached to the probe and detected by a labeled binding protein (typically an antibody) |
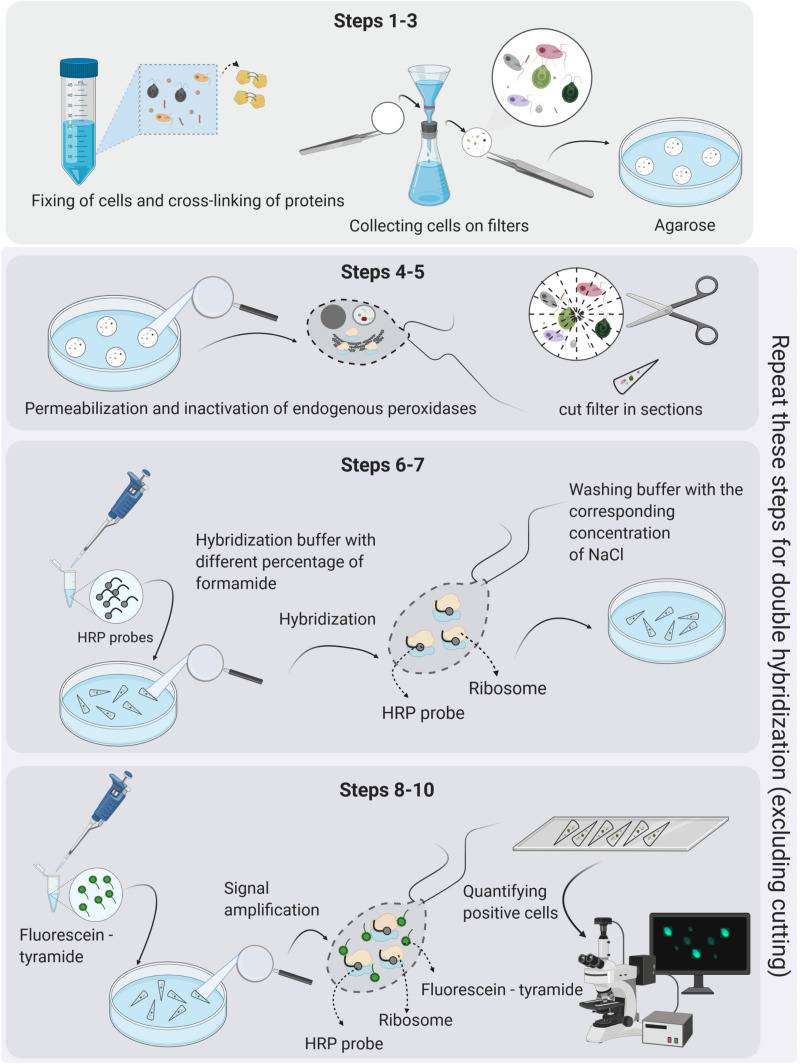
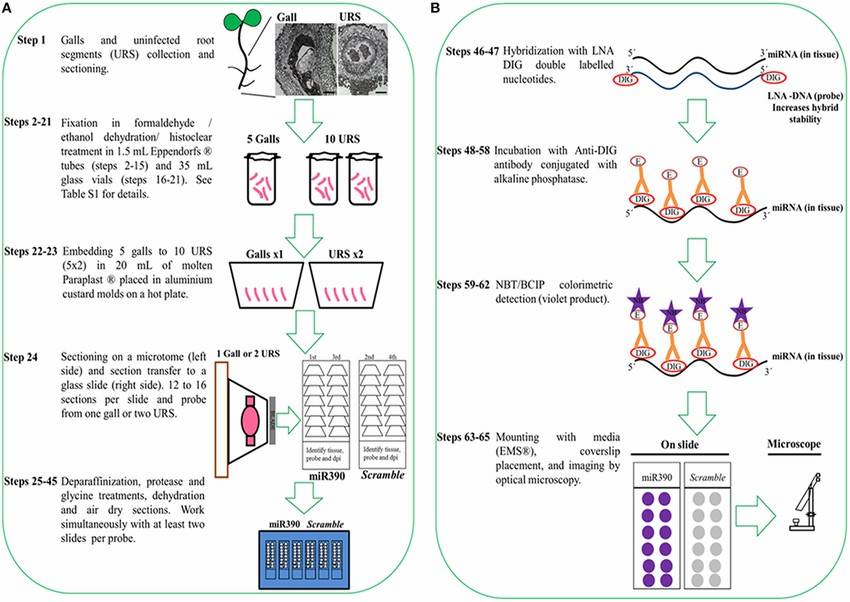 Fig. 3. Protocol flowchart. Schematic diagram of ISH procedure representing all necessary steps from sample collection to colorimetric detection of miRNAs (Díaz-Manzano FE, Barcala M, et al., 2016).
Fig. 3. Protocol flowchart. Schematic diagram of ISH procedure representing all necessary steps from sample collection to colorimetric detection of miRNAs (Díaz-Manzano FE, Barcala M, et al., 2016).
ISH Technology Optimization
Common problems and solutions
Table. 1. Troubleshooting guide for common problems in ISH (Jones J, Howat WJ. et al., 2020).
| Problem | Possible causes | Solutions |
| No signal |
|
|
| High background |
|
|
| High signal |
|
|
| Background in wrong cell/tissue types |
|
|
| Nuclear signal |
|
|
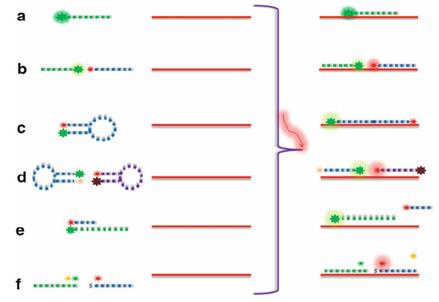
Signal amplification techniques
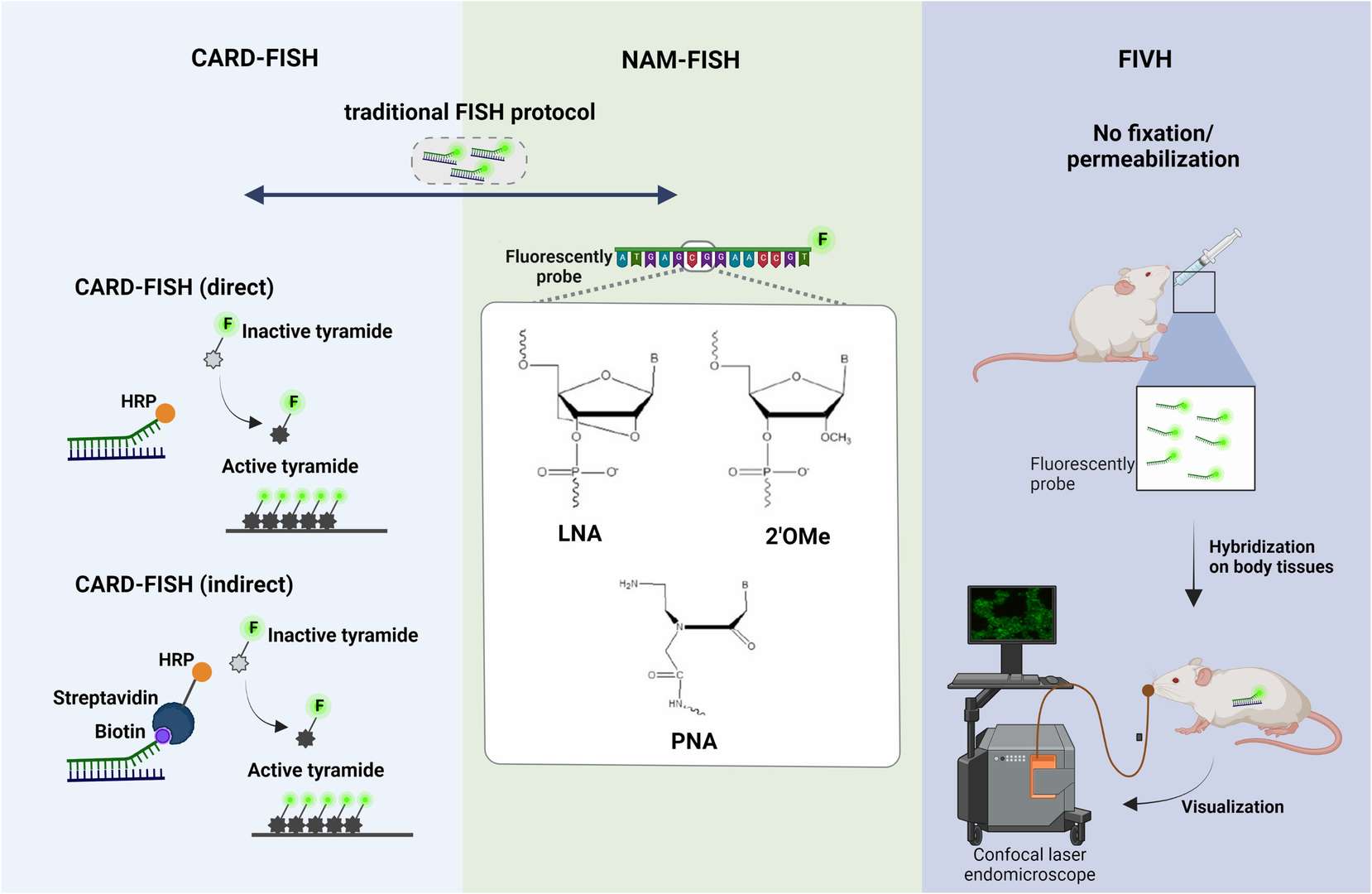
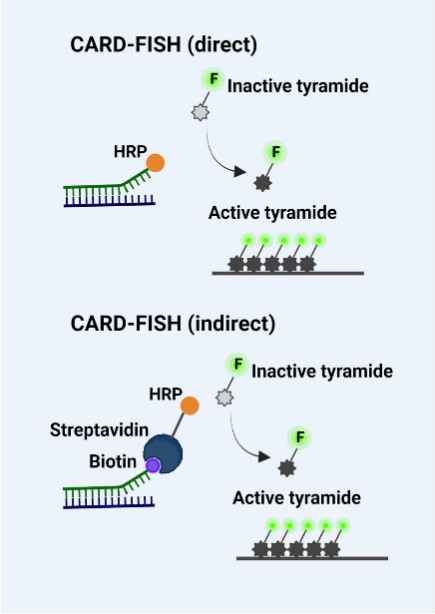
Tyramide signal amplification (TSA/CARD): Tyramine Signal Amplification (TSA/CARD): This procedure employs the color-enhancing catalytic reporter technique (CARD). It works by affixed hundreds of fluorescent tyramine molecules where the probe crosses paths with the target molecule under the catalytic action of horseradish peroxidase (HRP). The LNA (locked nucleic acid) probe is labelled with digitonin (DIG) and, upon hybridization with the miRNA target, can be targeted to the hybridization location by HRP-labeled anti-DIG antibody. By adding tyramine substrate, it produces an intense fluorescent light at the desired position, visible with fluorescence microscopy.
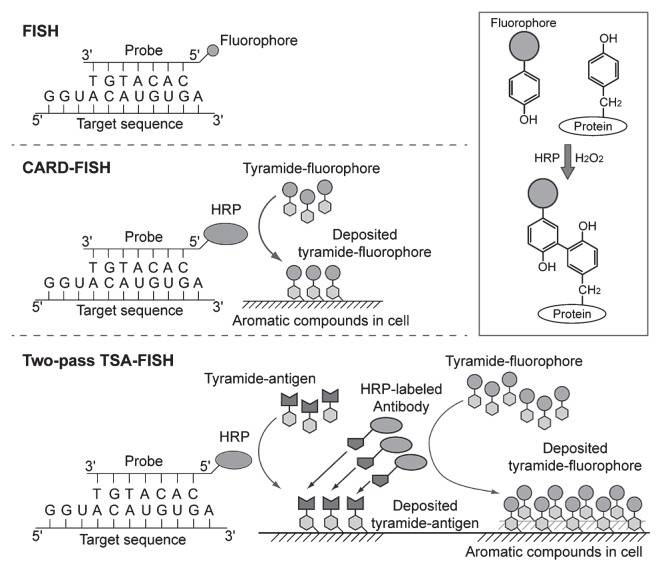 Fig. 4. Schematic depiction of FISH, CARD-FISH, and two-pass TSA-FISH. The scheme in the box depicts tyramide immobilization on tyrosine (Kubota K, 2013).
Fig. 4. Schematic depiction of FISH, CARD-FISH, and two-pass TSA-FISH. The scheme in the box depicts tyramide immobilization on tyrosine (Kubota K, 2013).
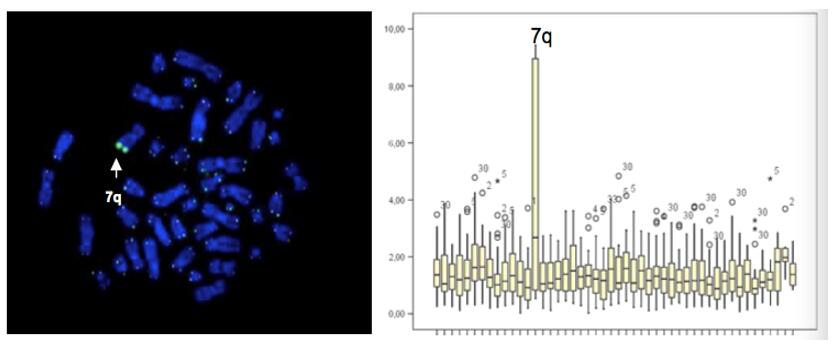
Rolling circle amplification (RCA): Following the technique of rolling circle replication, a circular DNA molecule is generated by hybridization with a padlock probe and the signal amplified by the DNA polymerase reaction. It produces an ultralong RCP that can provide multiple hybridization sites for enzyme- or fluorophore-labelled oligonucleotides at significant signal amplification. This approach can be used to identify single nucleotide polymorphisms (SNPs) and gene sequences.
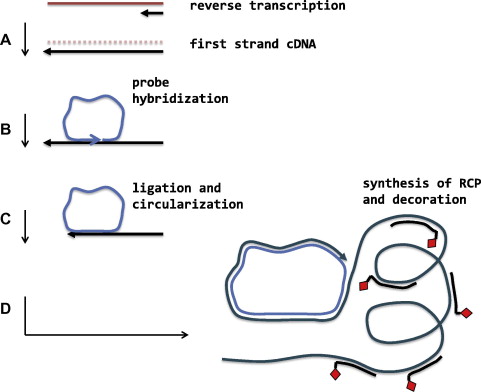 Fig. 5. Rolling circle amplification with padlock probe (Cassidy A, and Jones J, 2014).
Fig. 5. Rolling circle amplification with padlock probe (Cassidy A, and Jones J, 2014).
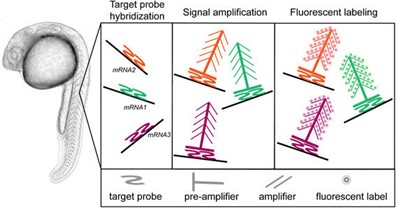
Branching DNA amplification (bDNA): signal amplification by creating a branching tree of complementary sequences coupled to targeted oligonucleotide probes. This approach relies on amplification of multiple signals by combining preamplifiers and amplifier chains. The method allows for extremely high amplification and therefore the assay is highly sensitive.
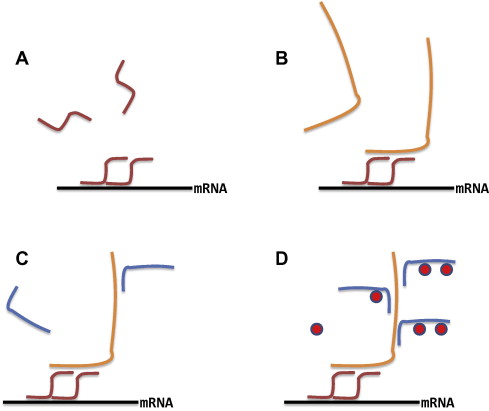 Fig. 6. Sequential hybridisations build up a branched structure in order to increase the number of binding sites for the AP conjugated oligonucleotide probe (Cassidy A, and Jones J, 2014).
Fig. 6. Sequential hybridisations build up a branched structure in order to increase the number of binding sites for the AP conjugated oligonucleotide probe (Cassidy A, and Jones J, 2014).
ISH Types
With the development of technology, there is a wide variety of in situ hybridization-derived techniques in addition to FISH. This section introduces several of the most commonly used ISH techniques.
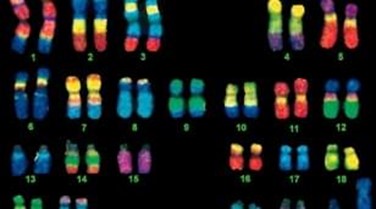
Multi-color FISH
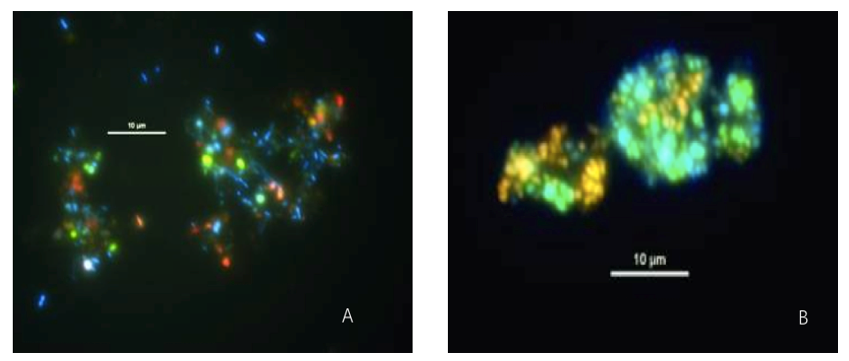
Multi-color FISH is a technique that produces multiple detection by individually labelling multiple probes. It can be used to measure multiple chromosomal loci at once (for example, advanced genomic editing) and is ideally suited to the analysis of multiple chromosome anomalies.
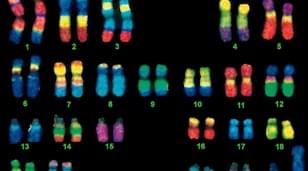
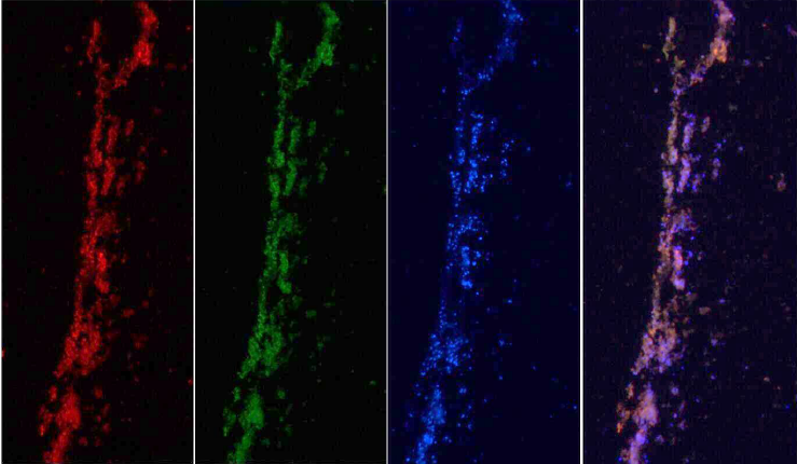
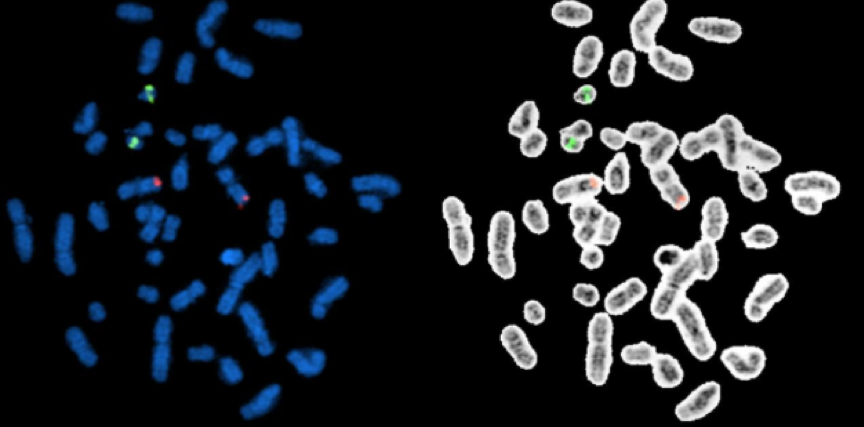
 Fig. 7. Simultaneous visualization, by multicolor FISH, of seven phylogenetically distinct microbial isolates in a mixture of pure cultures (mock community) (Lukumbuzya M, Schmid M, et al., 2019).
Fig. 7. Simultaneous visualization, by multicolor FISH, of seven phylogenetically distinct microbial isolates in a mixture of pure cultures (mock community) (Lukumbuzya M, Schmid M, et al., 2019).
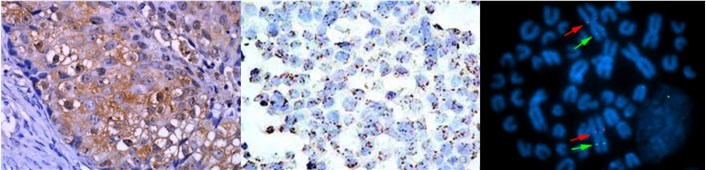
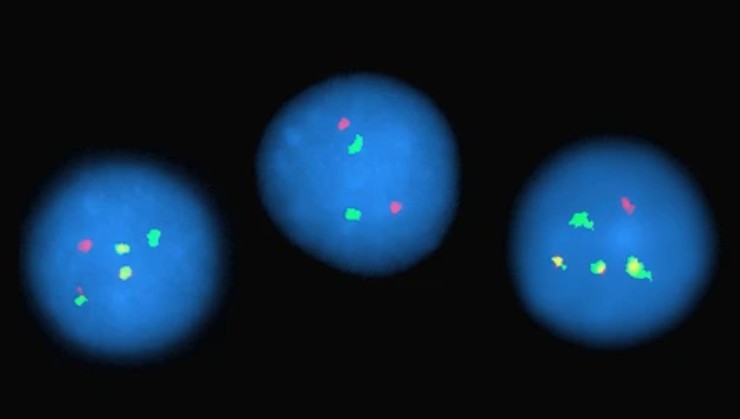
Chromogenic in situ hybridization (CISH)
CISH is a variant of in situ hybridisation that is widely used to identify gene amplifications, deletions, chromosomal translocations and quantitative changes. Unlike FISH, CISH involves an old-fashioned peroxidase or alkaline phosphatase reaction, which is portrayed using bright-field microscopy.
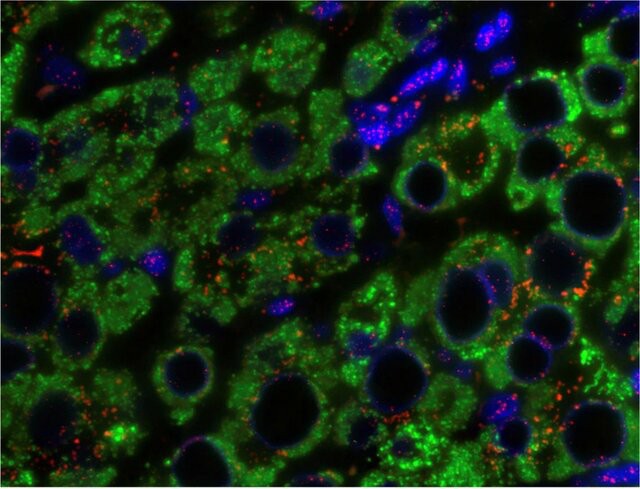
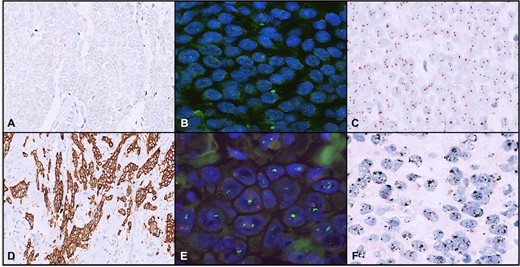
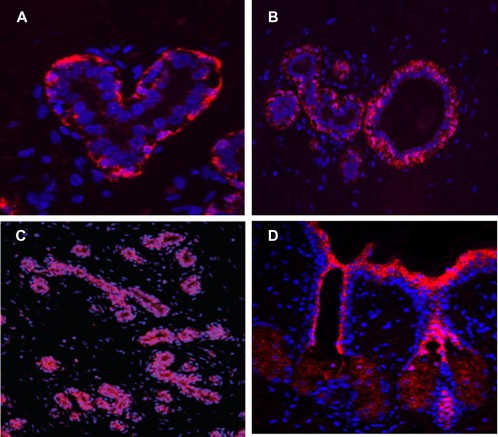
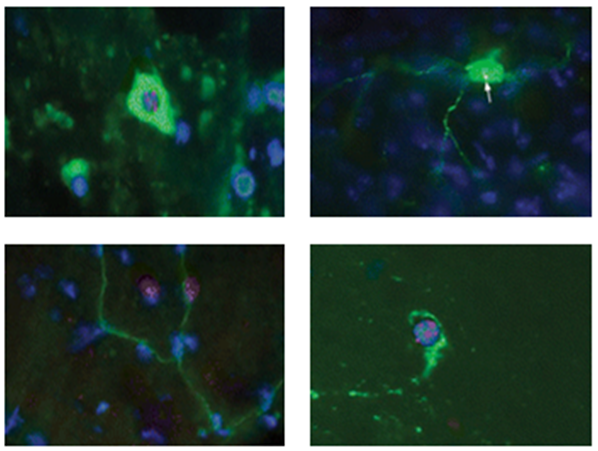
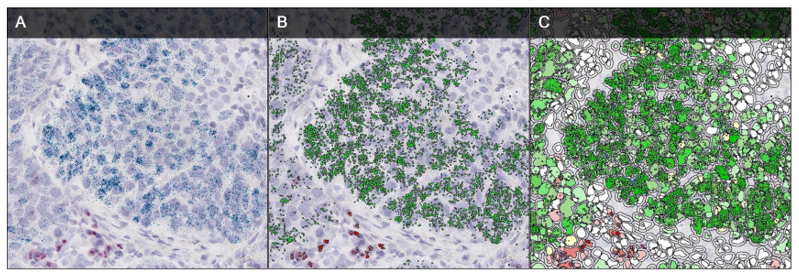
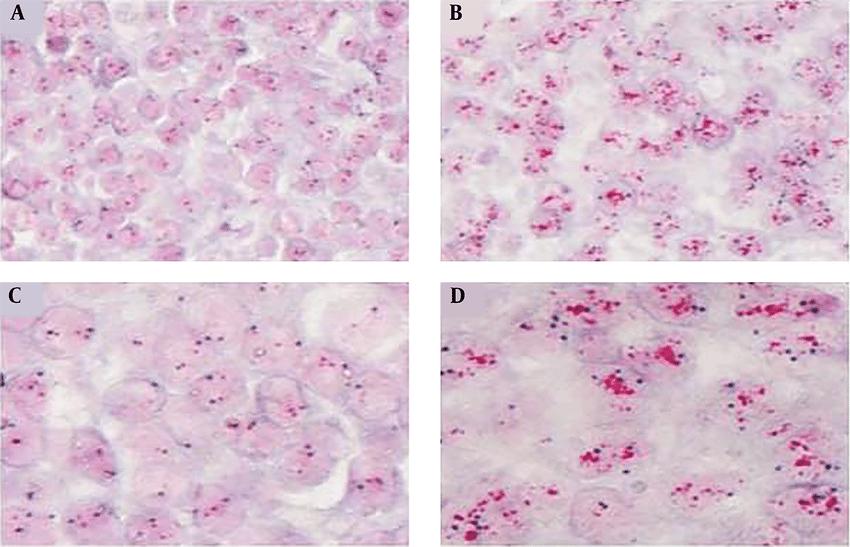 Fig. 8. An example of a chromogenic in situ hybridization of normal (A and C) and human epidermal growth factor receptor 2-positive breast cancer (B and D) cells (Esmaeili M, Mirzaahmadi S, et al., 2019).
Fig. 8. An example of a chromogenic in situ hybridization of normal (A and C) and human epidermal growth factor receptor 2-positive breast cancer (B and D) cells (Esmaeili M, Mirzaahmadi S, et al., 2019).
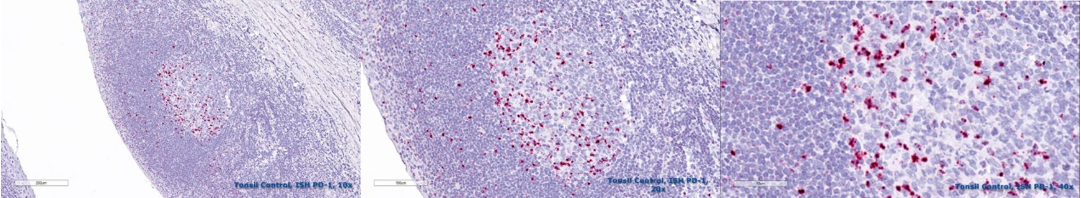
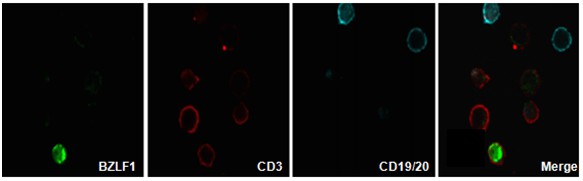
RNA in situ hybridization (RNA ISH)
RNA-ISH is an imaging procedure that detects the localisation of specific RNA molecules in tissues or cells. It can detect and localize transcription products (mRNA, non-coding RNA, etc.) directly. regulated expression of genes, and so has important uses in gene expression and transcription regulation.
Immunohistochemical in situ hybridization (Immuno-ISH)
Immuno-ISH combines the benefits of in situ hybridization and immunohistochemistry (IHC). By combining antibody detection with hybridization probes, Immuno-ISH can detect individual RNA molecules and associated proteins all at once.
Spectral in situ hybridization (SISH)
SISH combines FISH technology with spectroscopic analysis for more efficient chromosome multiplexing. By analyzing the fluorescence spectra of different chromosomes, it allows for more precise genomic analysis in a group of samples.
ISH Applications
Diagnostic histopathology
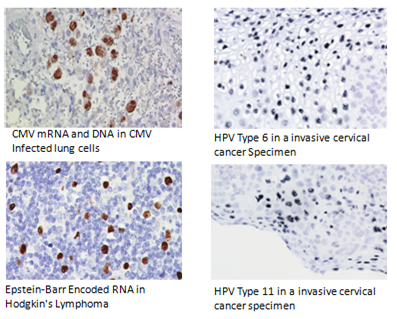
ISH is an excellent method for studying histopathology, particularly infectious disease and gene expression. ISH uses DNA probes to detect virus, bacteria and fungus genes within cells and tissues. This is vital in the diagnosis of infectious diseases. For instance, cytomegalovirus and herpesvirus can be detected using ISH in formalin-fixed paraffin-embedded sections.
Services: In Situ Detection of Virus
Cancer diagnosis
FISH technology has critical diagnostic and prognostic roles in cancer research. FISH can help with disease diagnosis by finding certain chromosome or gene abnormalities, like the Philadelphia chromosome in chronic granulocytic leukemia. It also identifies amplifications and deletions of cancer genes to inform treatment choices, for example, by assessing whether or not the HER2 gene is present in breast cancer.
Services: Circulating Tumor Cell (CTC) FISH, Neoplasms FISH Analysis
Gene expression studies
ISH is used to study gene expression in healthy and diseased tissues. It's effective for determining the regulation, storage and secretion of hormones, and ISH is especially useful for diagnosing neuroendocrine conditions when immunohistochemistry isn't a viable test.
Genetic studies
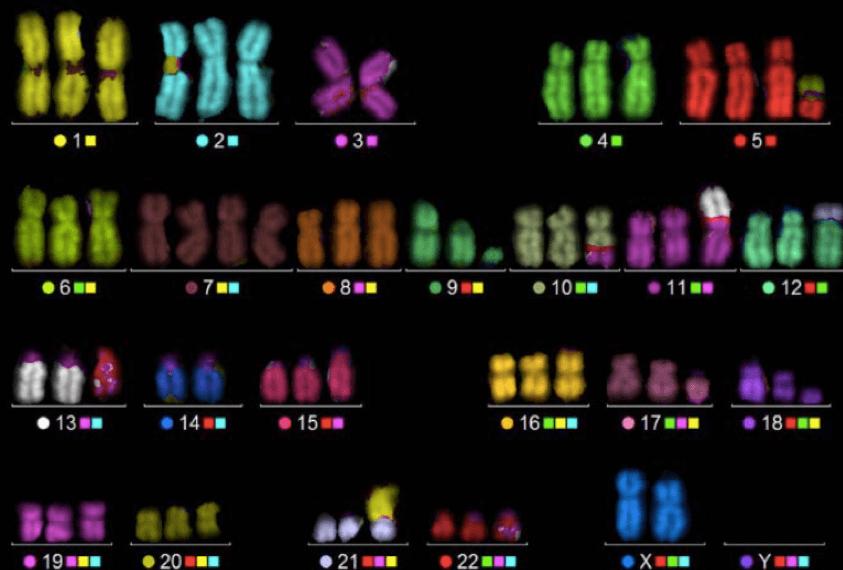
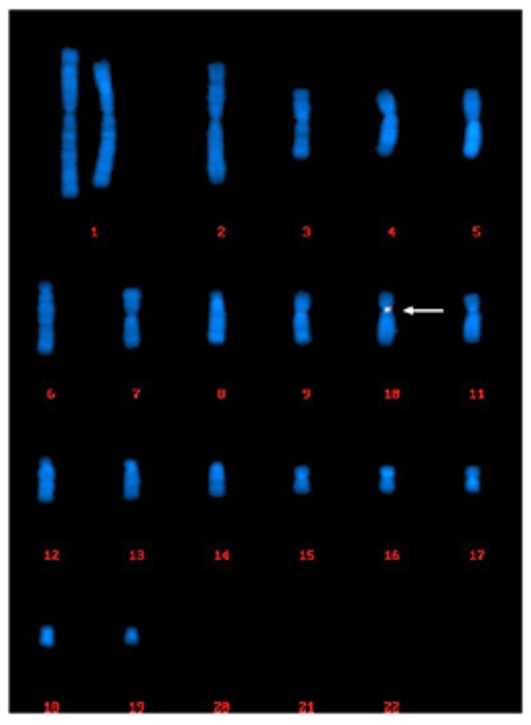
ISH, specifically FISH, is used to identify chromosomal abnormalities. It can be used for pre-diagnosis, like chromosome number mismatches such as Down syndrome. FISH is also used to localize genes and map chromosomes in order to create high resolution chromosome maps.
Services: Splice Variant Analysis (FISH), Telomere Length Analysis (Q-FISH)
Microbiology research
In microbial ecology, FISH is applied to the detection and localisation of certain groups of microbes. This enables us to detect and quantify various microbial communities in environmental samples, and optimize operations such as wastewater treatment.
Services: FISH Analysis of Microorganisms, CARD-FISH for Environmental Microorganisms
Exploration of temporal and spatial patterns of gene expression.
RNA-FISH allows us to measure gene expression in a single cell and compare gene expression in different cell types and conditions. This is relevant for the intricate web of gene regulation in, say, embryonic development.
References
- Ehtisham, M. Fundamentals of in situ hybridization: a review. 2016.
- Díaz-Manzano FE, Barcala M, Engler G, Fenoll C, de Almeida-Engler J, Escobar C. A Reliable Protocol for In situ microRNAs Detection in Feeding Sites Induced by Root-Knot Nematodes. Front Plant Sci. 2016. 7:966.
- Jones J, Howat WJ. Guidelines for the Optimization and Validation of In Situ Hybridization. Methods Mol Biol. 2020. 2148:3-17.
- Kubota K. CARD-FISH for environmental microorganisms: technical advancement and future applications. Microbes Environ. 2013. 28(1):3-12.
- Cassidy A, Jones J. Developments in in situ hybridisation. Methods. 2014. 70(1):39-45.
- Lukumbuzya M, Schmid M, et al. A Multicolor Fluorescence in situ Hybridization Approach Using an Extended Set of Fluorophores to Visualize Microorganisms. Front Microbiol. 2019. 19;10:1383.
- Kushkevych I, Hýžová B, et al. Microscopic Methods for Identification of Sulfate-Reducing Bacteria from Various Habitats. Int J Mol Sci. 2021. 13;22(8):4007.
- Esmaeili M, Mirzaahmadi S, et al. Evaluation of additional abcc12 gene expression character in breast cancer samples using formal diagnostic profile. Gene Cell and Tissue, In Press. 2019.