Overview of Oligo-FISH Technology
Fluorescence in situ hybridisation (FISH) has been a revolutionary method in molecular genetics since its development in 1982. Classic FISH uses giant genomic insertions such as bacterial artificial chromosomes (BACs) and repeating DNA sequences as probes. These standard probes, although they work well, are limited in chromosomal and chromosome fragment recognition. A significant alternative, oligonucleotide-based fluorescent in situ hybridisation (Oligo-FISH), uses computationally screened and engineered oligonucleotide probes to overcome these restrictions. Oligo-FISH has the advantage of being more molecularly specific and sensitive - an advantage that makes it especially useful for research with highly heterogeneous plant chromosomes. Not only does this increase experimental reproducibility and stability, but also lowers resource consumption, which opens up vast opportunities for plant cytogenetics.
What is Oligo-FISH Technology?
Oligo-FISH, or oligonucleotide-based fluorescence in situ hybridisation, uses computationally selected and engineered oligonucleotide probes fluorophore-labelled to locate DNA sequences. The key to Oligo-FISH is to create and manufacture these oligonucleotide probes (usually 20-50 nucleotides long). These probes are designed specifically to attach to chromosomes or chromosome regions.
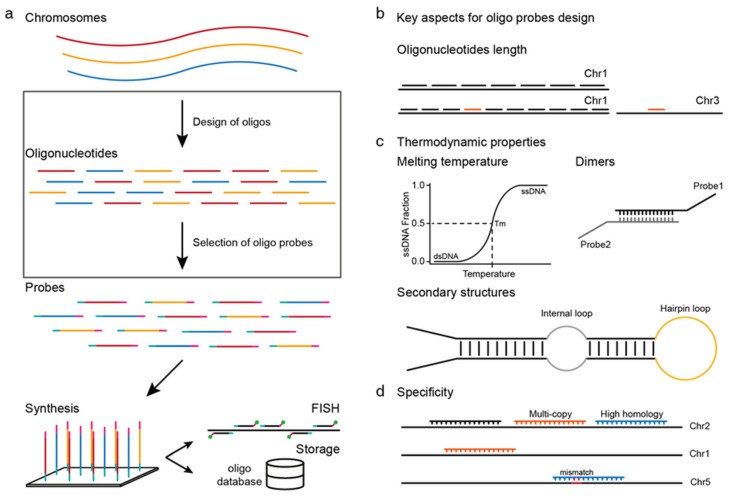 Fig. 1. Workflow and the key aspects of oligonucleotide fluorescence in situ hybridization (oligo-FISH) probe design (Liu G, Zhang T, et al., 2021).
Fig. 1. Workflow and the key aspects of oligonucleotide fluorescence in situ hybridization (oligo-FISH) probe design (Liu G, Zhang T, et al., 2021).
Oligonucleotide probes preparation:
Oligonucleotide probes are designed using computation: dedicated software (such as Chorus2) transcribes target chromosome or genome sequences into short oligonucleotide fragments. The fragments are then tested for highly specialized single-copy oligonucleotide sequences on the basis of melting temperature, sequence complexity, secondary structure prediction and dimer formation prediction. The selected probes are synthesised simultaneously using a synthesizer and then labelled with biotin, digoxigenin or fluorescent probes. Oligonucleotide probes generally fall into one of two categories:
- Single-copy sequence oligonucleotide probes: These probes, based on single-copy DNA sequences, show high specificity and are used in studies of specific chromosomal fragments or entire chromosomes, including investigations of chromosomal evolution and meiotic pairing. However, their application is limited in the absence of comprehensive genomic information.
- Repetitive sequence oligonucleotide probes: Designed from microsatellite repeats, satellite sequences, and tandem repeats, these probes do not depend on genome sequencing information. They are suitable for karyotype analysis, phylogenetic studies, and nondisjunction FISH (ND-FISH).
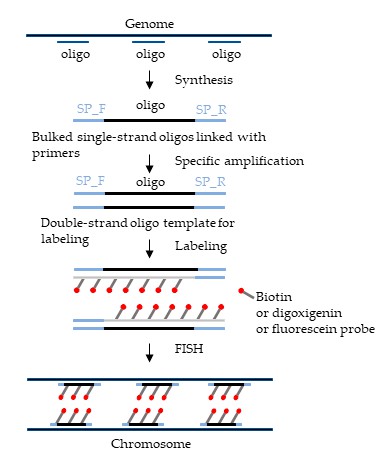 Fig. 2. The schematic diagram represents oligo probe library enrichment, PCR amplification for primer binding, fluorophore labeling for de novo probe synthesis, and FISH experiment being performed with developed oligo probes (Harun A, Liu H, et al., 2023).
Fig. 2. The schematic diagram represents oligo probe library enrichment, PCR amplification for primer binding, fluorophore labeling for de novo probe synthesis, and FISH experiment being performed with developed oligo probes (Harun A, Liu H, et al., 2023).
Key aspects of oligonucleotide probe preparation:
Designing high-quality single-copy oligonucleotide probes is paramount for successful Oligo-FISH experiments. Several factors must be considered:
- Minimizing repetitive sequences in target sequences: Utilize tools like BLAST, Bowtie2, and BWA to filter out repetitive sequences, enhancing probe specificity.
- Probe density: Test different densities of oligonucleotide probes to determine optimal signal intensity. Typically, 0.1-0.5 oligos/kb density suits condensed metaphase chromosomes, whereas higher densities (e.g., 2 oligos/kb) are required for pachytene chromosomes.
- Probe length: Select appropriate oligonucleotide lengths considering specificity, stability, and binding efficiency. While longer probes present higher specificity, they may form secondary structures or hairpins, reducing hybridization efficiency. Shorter probes generally yield higher FISH signals.
- Thermodynamic properties: Accurately estimate the probe's Tm, considering GC content, salt concentration, and nearest-neighbor interactions to ensure optimal hybridization temperatures.
- Avoiding secondary structure and dimers: Carefully design probes to prevent folding into secondary structures (e.g., hairpins) that reduce hybridization efficiency. Assess salt concentrations and nearest-neighbor parameters to mitigate dimer formation.
- Specificity design: Select high-quality reference genomes covering most repetitive elements and utilize de novo repeat detection and filtering methods to enhance specificity.
Advantages and Limitations of Oligo-FISH Technology
Advantages:
- Enhanced specificity and sensitivity: Computationally selected probes exhibit high specificity, effectively reducing background noise and enhancing signal sensitivity.
- Flexible design: Oligonucleotide probes can be flexibly tailored to detect both single-copy and repetitive sequences, catering to diverse experimental needs.
- Library usage: Short oligonucleotide probes are easily synthesized, amplified, and labeled, making them suitable as probe libraries.
- Multiplexing capability: Capable of detecting multiple DNA targets in a single assay, facilitating complex chromosomal studies.
- Higher resolution: Provides superior spatial resolution, suitable for single-cell-level gene expression and chromosomal structure studies.
Limitations:
- Technical complexity: Designing, synthesizing, and labeling oligonucleotide probes require significant technical expertise, high-throughput sequencing, and computational tools.
- Stringent experimental conditions: The experimental process is intricate and conditions must be rigorously maintained, with slight deviations potentially impacting the results.
- Limited application scope: Complex genome structures and large chromosomes still pose detection challenges, and probe design is constrained by the completeness of genomic information, limiting studies in species without comprehensive genome sequences.
Applications of Oligo-FISH in Plant Genomic Research
Oligo-FISH significantly enhances the precision and efficiency of karyotype analysis in plant genomic research, playing a vital role in studying genome structure and function. By using specific probes, researchers can conduct detailed karyotype analysis across different species, aiding in understanding plant chromosomal evolution and providing technical support for non-model plant chromosome identification.
- Chromosome identification and karyotype analysis: Utilize probes targeting chromosome arms, fragments, and centromeric sequences for chromosome identification in various plants like oranges, cucumbers, and maize.
- Chromosomal rearrangement and fusion studies: Employ low-copy oligonucleotide probes to identify chromosomal translocations in plants. For instance, identifying specific chromosomal translocations in rice and wheat cultivars supports genetic linkage map construction.
- Polyploid plant research: Oligo-FISH provides an essential platform for studying genome and evolutionary analyses in polyploid plants, revealing mechanisms of homoeologous and non-homoeologous chromosome pairing in complex genomes.
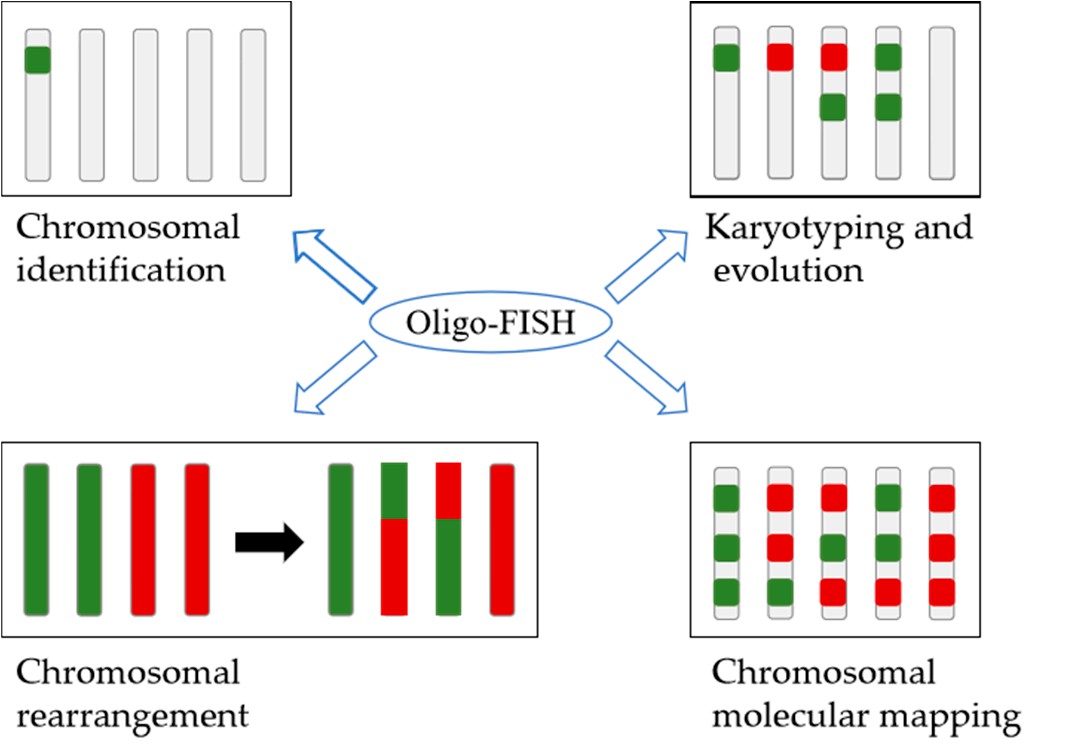 Fig. 3. Applications of oligo-FISH in plants (Harun A, Liu H, et al., 2023).
Fig. 3. Applications of oligo-FISH in plants (Harun A, Liu H, et al., 2023).
Oligo-FISH technology offers revolutionary advancements in molecular genetics and plant cytogenetics, filling gaps left by traditional FISH techniques. Its high specificity, flexibility, and resolution make it a powerful tool for chromosomal research and functional genomics. Addressing the limitations and challenges through continuous advancements will further broaden its applications, ultimately enhancing our understanding of plant genetics and fostering significant improvements in crop breeding and genetic research.
| Products & Services | Description |
| Fluorescent In Situ Hbridization (FISH) Service | Creative Bioarray offers a range of different FISH services including metaphase and interphase FISH, fibre-FISH, RNA-FISH, M-FISH, 3D-FISH, flow-FISH, FISH on paraffin sections, and immune-FISH. |
| FISH Probe Design, Synthesis, and Testing Service | Creative Bioarray is capable of developing custom FISH probes. Apart from that, we also offer mRNA ISH/FISH probes, miRNA ISH/FISH probes, and lncRNA ISH/FISH probes. |
| FISH Quality Control Services | Creative Bioarray has years of experience in performing FISH, and cytogenetic services, we can also provide custom FISH quality control service for your lab's needs! |
References
- Liu G, et al. Single Copy Oligonucleotide Fluorescence In Situ Hybridization Probe Design Platforms: Development, Application and Evaluation. Int J Mol Sci. 2021. 1;22(13):7124.
- Harun A, et al. Oligonucleotide Fluorescence In Situ Hybridization: An Efficient Chromosome Painting Method in Plants. Plants. 2023. 12(15):2816.

