What are the Differences between FISH, aCGH, and NGS?
In the modern medicine, genetic testing technologies play a pivotal role. They allow scientists and clinicians to examine thoroughly in disease-related biomarkers and genetic variants, giving patients more accurate diagnoses and treatment options. This article will introduce you to three major genetic testing technologies - fluorescence in situ hybridization (FISH), array comparative genomic hybridisation (aCGH), and next-generation sequencing (NGS). We'll talk about them individually in terms of their underlying principle, technical features, application cases, and strengths and weaknesses.
FISH Technology
FISH is one of the most common molecular cytogenetic techniques. It uses fluorescent probes to amplify DNA or RNA sequences in chromosomes or cells. With fluorescence microscopy, the probe's position can be read visually and used to determine the position and copy number of a given genetic sequence. FISH technology is all about designing probes and fluorescent labeling (either with direct or indirect fluorescent dyes). Probes hybridize with target sequences, and their hybridisation patterns are measured with a fluorescence microscope.
FISH technology finds applications across genetic counselling, cancer testing and research, cytology, pathology, and species identification. For example, it detects the BCR/ABL translocation in leukemia cells to confirm the diagnosis of chronic myelogenous leukemia.
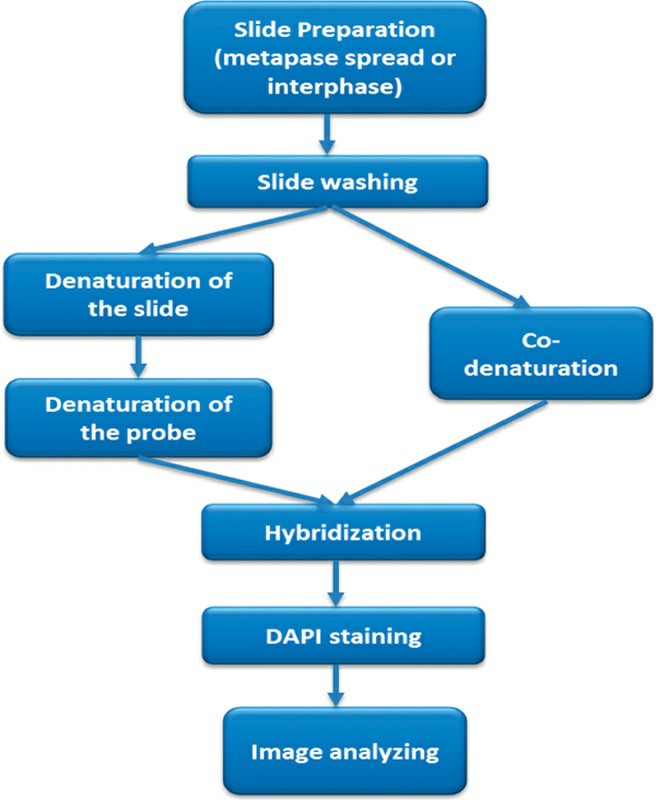 Fig. 1. Workflow of the FISH technique (Eid, Ola M., 2017).
Fig. 1. Workflow of the FISH technique (Eid, Ola M., 2017).
Advantages:
- High resolution: Capable of detecting fine genetic mutations and chromosomal anomalies.
- Rapid turnaround: Suitable for rapid diagnoses, such as prenatal screening.
- High compatibility: Can be combined with other detection methods like PCR and microarrays.
- Visualization: Enables direct visualization of detection results via fluorescence microscopy.
Limitations:
- Target limitation: Limited to detecting known sequences or specific probe-designed regions.
- Balanced rearrangements: May not detect balanced translocations and inversions.
aCGH Technology
aCGH is a high-throughput method for identifying copy number variations (CNVs) in genomic DNA. This method involves comparing fluorescent signal intensities between sample DNA and reference DNA to identify potential copy number increases or decreases within the genome. aCGH leverages DNA microarrays paired with probes from several chromosomes to detect genomic changes in a highly accurate way.
aCGH has been used extensively in cancer research, genomic abnormalities (microdeletion and duplication syndromes), and prenatal genetic testing. It is used, for instance, to map microdeletion spots in hereditary syndromes or to measure copy number changes in tumours.
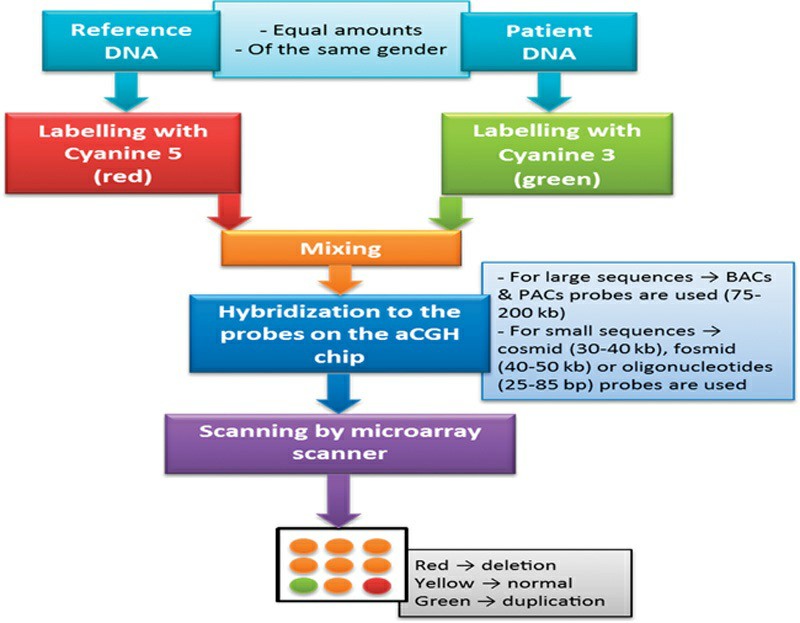 Fig. 2. Workflow of aCGH technique (Eid, Ola M., 2017).
Fig. 2. Workflow of aCGH technique (Eid, Ola M., 2017).
Advantages:
- Broad detection range: Capable of identifying most CNVs across the genome.
- High resolution: Detects variations as small as single exons.
- High throughput: Capability to analyze numerous genomic regions simultaneously.
- Automated data processing: Simplifies result visualization and interpretation.
- Direct application: Can directly analyze clinical samples without the need for cell culture.
Limitations:
- Copy-neutral variations: Cannot detect balanced genetic variations like translocations and inversions.
- Sample quality: Requires high-quality DNA samples.
- Complex interpretation: May face complexities and potential false positives in result interpretation.
NGS Technology
NGS, or second-generation sequencing, refers to a variety of high-throughput DNA sequencing techniques. NGS parallelises and miniaturizes sequencing machines to create enormous amounts of sequence data in a single run. There are several steps involved: collecting the samples, sequencing the reactions, and data analysis. NGS uses a variety of techniques, including synthesis-based sequencing, fluorescently labeled nucleotides, and real-time sequencing.
NGS is widely used for whole-genome sequencing, exome sequencing, transcriptomics, epigenetic studies, and diagnostics. For example, it identifies enigmatic disease-related variants in genetic disorders.
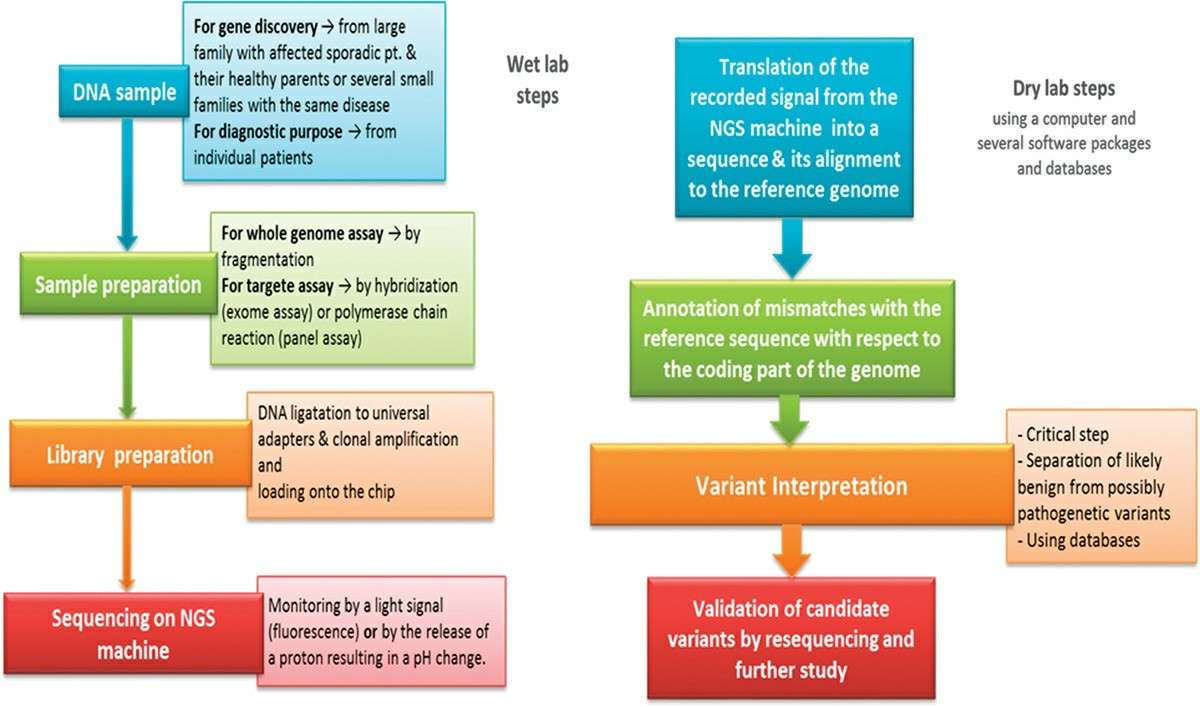 Fig. 3. Workflow of a NGS analysis (Eid, Ola M., 2017).
Fig. 3. Workflow of a NGS analysis (Eid, Ola M., 2017).
Advantages:
- High coverage and resolution: Achieves deep sequencing at high resolution.
- Multivariate detection: Simultaneously detects various types of genetic variations.
- Scalable research: Ideal for large-scale studies, like Genome-Wide Association Studies (GWAS).
Limitations:
- High cost and resource intensive: Demands significant financial and computational resources.
- Complex data handling: Involves complex data processing and storage.
- Bioinformatics expertise: Requires extensive bioinformatics analysis and specialized knowledge.
Comparison of FISH, aCGH, and NGS
| FISH | aCGH | NGS | |
| Detection range | Targets single or few known genes or sequences. | Genome-wide detection of copy number variations. | Broad scope, suitable from single gene to entire genome scales. |
| Accuracy | High accuracy but limited range. | High resolution but unable to detect balanced variations. | Extremely high accuracy and resolution, covers a wide range of variant types. |
| Detection Time | Moderate, suitable for rapid diagnosis. | Faster, ideal for high-throughput screening. | Relatively slower but comprehensive. |
| Application Scenarios | Diagnosing known chromosomal abnormalities, targeted cancer and genetic disease detection. | Comprehensive genomic variation analysis, cancer research, genetic disease screening. | Whole-genome analysis, genetic research of complex diseases, personalized medicine. |
Each of these technologies - FISH, aCGH, NGS - offers its own advantages and limitations. Depending on the purpose of a study or diagnostic procedure, one technology might work better than another. FISH delivers accurate and rapid insights, aCGH delivers CNVs at the macro level, and NGS delivers massive and very detailed genomic data. Knowing these differences allows one to decide the best approach for a particular medical or research use.
| Products & Services | Description |
| Fluorescent In Situ Hbridization (FISH) Service | Creative Bioarray offers a range of different FISH services including metaphase and interphase FISH, fibre-FISH, RNA-FISH, M-FISH, 3D-FISH, flow-FISH, FISH on paraffin sections, and immune-FISH. |
| Molecular Karyotyping (aCGH) Service | Creative Bioarray offers a wide range of Karyotyping Services (Traditional Karyotyping-G Banded, M-FISH, Molecular Karyotyping, etc) to meet your different needs. Creative Bioarray can provide you with the best research support in karyotyping analysis field. |
| FISH-Based Clonality Analysis Services | Creative Bioarray offers comprehensive fluorescence in situ hybridization (FISH)-based clonality analysis services to support our clients in maintaining the highest standards of product quality and regulatory compliance. |
References
- Eid, Ola M. Molecular cytogenetic techniques for identification of copy-number variations[J]. Middle East Journal of Medical Genetics, 2017, 6(1). 1-12.
- Ikbal Atli E, et al. Pros and Cons for Fluorescent in Situ Hybridization, Karyotyping and Next Generation Sequencing for Diagnosis and Follow-up of Multiple Myeloma. Balkan J Med Genet. 2021. 23(2). 59-64.
- Szuhai K, et al. Microarray Techniques to Analyze Copy-Number Alterations in Genomic DNA: Array Comparative Genomic Hybridization and Single-Nucleotide Polymorphism Array. J Invest Dermatol. 2015. 135(10). e37.

