What are Single, Dual, and Multiplex ISH?
In Situ Hybridization (ISH) is used in molecular biology and biomedical science to detect and identify specific nucleic acid molecules (DNA or RNA) in cells or tissues. ISH has continued to progress from Single In Situ Hybridization (single ISH) through Dual ISH and now more sophisticated Multiple In Situ Hybridisation (MISH) - where researchers can identify multiple target sequences within a single sample. Such techniques are outlined briefly in this article.
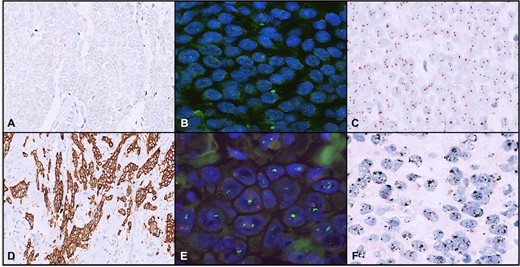 Fig. 1. A case with HER2/neu immunohistochemistry (IHC) score 1+ (A), nonamplified on both FISH (B) and dual-color dual in situ hybridization (D-DISH) (C). A case with HER2/neu IHC score 3+ (D), amplified on both FISH (E) and D-DISH (F) (Rathi, A., Sahay, A., et al., 2024).
Fig. 1. A case with HER2/neu immunohistochemistry (IHC) score 1+ (A), nonamplified on both FISH (B) and dual-color dual in situ hybridization (D-DISH) (C). A case with HER2/neu IHC score 3+ (D), amplified on both FISH (E) and D-DISH (F) (Rathi, A., Sahay, A., et al., 2024).
Single ISH
Single ISH is the process of examining the local presence of single nucleic acid fragments (DNA or RNA) in a cell or tissue sample. A method for identifying the locations of these sequences in a cell or tissue by hybridising with similar nucleic acid sequences in the cell/tissue using a radioactive, fluorescent or enzyme probe. It's an extensively used method for gene expression studies, disease surveillance, and virus identification.
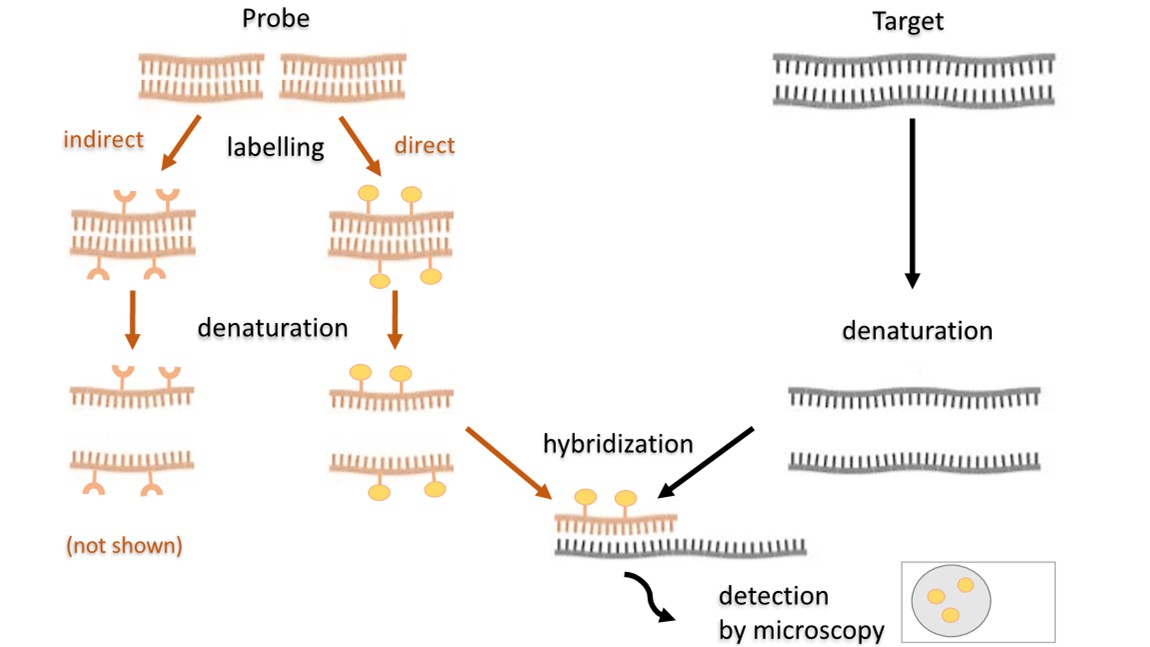 Fig. 2. Scheme of the fluorescence in situ hybridization (Kushkevych, I., Hýžová, B., et al., 2021).
Fig. 2. Scheme of the fluorescence in situ hybridization (Kushkevych, I., Hýžová, B., et al., 2021).
Features
- High specificity: Probes designed for single ISH are highly specific, enabling precise detection of target nucleic acid sequences.
- Accurate localization: Capable of directly visualizing the spatial distribution of target sequences within cells or tissues.
- Wide applicability: Suitable for various sample types, including fixed cells and tissue sections.
Applications
- Gene expression research: Single ISH is used for studying the expression of specific genes in different cells and tissues. For instance, it can be used to identify the epitopes of oncogenes in cancer research.
- Disease diagnosis: This is a technology that can detect viral infections by recognizing the virus RNA or DNA in host tissues, thereby diagnosing and predicting viral diseases.
- Developmental Biology: Single ISH allows us to understand embryonic development by exposing the spatiotemporal distribution of developmental genes.
Dual ISH
Dual ISH extends single ISH by allowing the simultaneous detection of two different nucleic acid sequences within the same cell or tissue section. This is achieved by using two differentially labeled probes, which can simultaneously provide more comprehensive research information.
Methods:
- Radionuclide and non-radioactive labeled probes: Common radioactive isotopes such as 35S and non-radioactive labels like biotin and digoxigenin are used. These methods can be divided into one-step and two-step procedures. The two-step reaction involves reacting two probes together, the signal detection to integrate with the chromogenic sample for non-radiolabeled target nucleic acids, the sample is dried, and radiolabelled autoradiography to display another target nucleic acid. The two-step approach, in which radiolabels are labelled and developed first, and biotin or digoxin labeled and developed, is time-consuming but the labelling signal is preserved.
- Non-radioisotope labeled probes: This method simultaneously employs biotin and digoxigenin for dual hybridization, followed by sequential color development using horseradish peroxidase-conjugated streptavidin for red detection and alkaline phosphatase-conjugated anti-digoxigenin antibody for blue detection. Another method is dual fluorescent labeling, which involves using different colored fluorophores to label different target nucleic acids. These can be directly observed under a fluorescence microscope by selecting different excitation filters.
- Two radioisotope labeled probes: Probes labeled with 3H and 35S are mixed for concurrent hybridization. During autoradiography, the sample is covered with two layers of nuclear emulsion separated by a transparent plastic film. Low-energy beta rays from 3H signal the first emulsion layer, while high-energy beta rays from 35S penetrate the plastic film to expose the second layer, showing signals in both emulsions.
Features
- Dual detection: Capable of simultaneously detecting two target nucleic acid sequences within the same sample.
- Color coding: Different fluorescent or colorimetric markers distinguish various target sequences.
- High information content: Provides more comprehensive information on gene expression or disease-related nucleic acid sequences.
Applications
- Gene co-expression studies: By detecting the expression of two related genes simultaneously in cells or tissues, researchers can study their interactions and regulatory relationships. For instance, it is used in cancer research to detect the expression of oncogenes and tumor suppressor genes concurrently.
- Cell subgroup analysis: In immunological studies, dual ISH can differentiate gene expression differences among various cell subgroups, aiding in the understanding of immune system functions and regulatory mechanisms.
- Neuroscience research: Dual ISH is employed to study the co-expression of different neurotransmitter receptors in neurons, revealing neuronal functions and interactions.
Multiplexed ISH
With advances in technology, multiplexed ISH can be used to dramatically increase a study's information resource by exposing multiple probes simultaneously to several sequences. RNAscope and multicolor FISH are the most widely employed multiplexed ISH methods RNAscope technology involves a sequence of double Z probes, divided into upper and lower complementary sections (Z sections), which bind to a region of high sensitivity and specificity, so that only when both Z sections are bonded to the target RNA at the same time will the signal be amplified. RNA, signals are amplified only when both Z-parts bind to the target RNA, and the method allows multiple single-molecule RNAs to be detected and identified in the same cell at the same time. Multicolor FISH (e.g., 24 different colored probes) are employed for spectral karyotyping to measure changes within the genome.
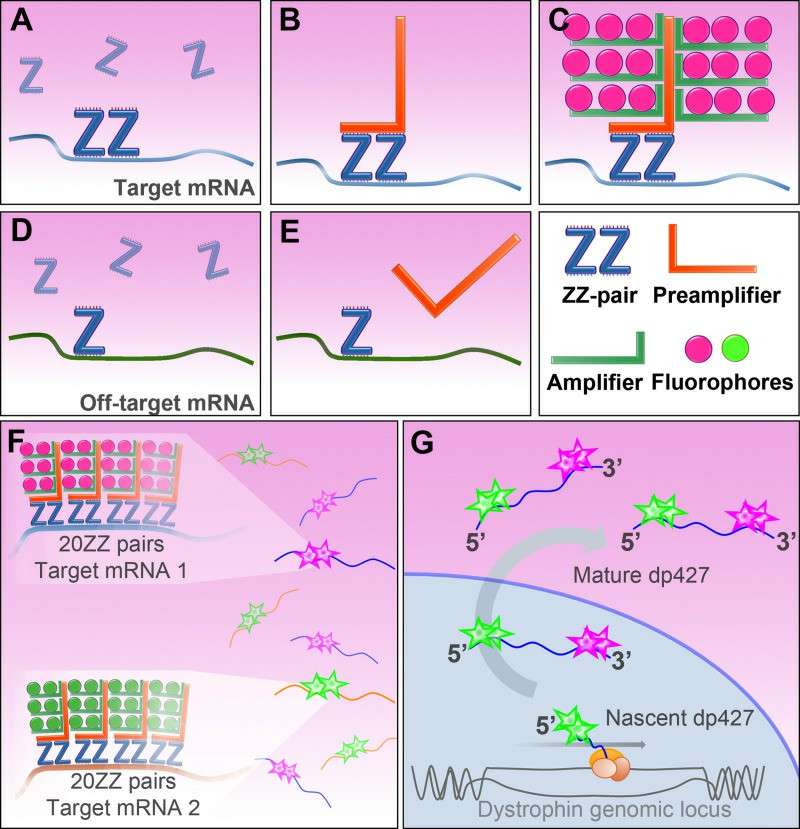 Fig. 3. RNAscope ISH method (Hildyard JCW, Rawson F, et al., 2020).
Fig. 3. RNAscope ISH method (Hildyard JCW, Rawson F, et al., 2020).
Applications
- Genomics research: In genomics, multiple ISH is utilized to map genes, identify gene expression patterns, and determine genome evolution.
- For Disease Research: In studies of complex diseases, like cancer and autoimmune diseases, multiple ISH devices can simultaneously detect multiple disease genes or molecular markers to generate the complete diagnostic and treatment data.
Single and dual ISH are fundamental to molecular biology and biomedical research. Single ISH has become an established technique for gene expression profiling and disease diagnosis because of its sensitivity and localization precision. Dual ISH offers more depth with multiplexed detection, and is especially well suited for studying gene interactions and patterns of gene expression.
| Products & Services | Description |
| Fluorescent In Situ Hbridization (FISH) Service | Creative Bioarray offers a range of different FISH services including metaphase and interphase FISH, fibre-FISH, RNA-FISH, M-FISH, 3D-FISH, flow-FISH, FISH on paraffin sections, and immune-FISH. |
| FISH Probe Design, Synthesis, and Testing Service | Creative Bioarray is capable of developing custom FISH probes. Apart from that, we also offer mRNA ISH/FISH probes, miRNA ISH/FISH probes, and lncRNA ISH/FISH probes. |
| FISH Quality Control Services | Creative Bioarray has years of experience in performing FISH, and cytogenetic services, we can also provide custom FISH quality control service for your lab's needs! |
References
- Rathi, A., Sahay, A., et al. Validation of Dual-Color Dual In Situ Hybridization for HER2/neu Gene in Breast Cancer. Archives of pathology & laboratory medicine, 2024, 148(4), 453–460.
- Kushkevych, I., et al. Microscopic Methods for Identification of Sulfate-Reducing Bacteria from Various Habitats. International journal of molecular sciences, 2021. 22(8), 4007.
- Hildyard JCW, Rawson F, et al. Multiplex in situ hybridization within a single transcript: RNAscope reveals dystrophin mRNA dynamics. PLoS One. 2020, 24;15(9), e0239467.

