Differences Between DNA and RNA Probes
In molecular biology and biotechnology, the probe now plays a crucial role in the recognition and sequencing of particular nucleic acid sequences. Two probes, DNA probes and RNA probes, are fundamentally different in structure, function and application. In this article, we will outline the major differences between DNA and RNA probes in in situ hybridization (ISH) as reference materials for molecular biologists.
Differences in Chemical Structure
- DNA probes: DNA probes generally consist of deoxyribonucleic acid (DNA) composed of four deoxynucleotide bases (adenine, thymine, cytosine, guanine). The bases link together by phosphodiester bonds to create a single strand. They are usually between 20 and 1000 bp, and some FISH probes reach 1-10 Kb depending on experimental requirements.
- RNA probes: RNA probes, by contrast, are made of ribonucleic acid (RNA) and comprise of four ribonucleotide bases (adenine, uracil, cytosine, guanine). Because of the 2' hydroxyl group in RNA, it is more chemically unstable and easily hydrolysed than DNA.
Synthesis and Labeling Methods
DNA probes
- Chemical synthesis: Stepwise synthesis of nucleotide sequences to produce highly pure DNA probes.
- Polymerase chain reaction (PCR): Amplification of specific DNA sequences using primers to generate desired probes.
Common labeling methods include fluorescent dyes, radioactive isotopes, chemiluminescence, enzyme labels, and biotin.
RNA probes
- In vitro transcription (IVT): Synthesis of RNA probes using DNA templates and RNA polymerase.
- Enzymatic modifications: Achieving high specificity and fidelity of RNA probes through specific enzymatic modifications.
Labeling typically includes incorporating labeled nucleotides during transcription, with common labels being fluorescent dyes and radioactive isotopes.
Classification of DNA and RNA Probes
DNA probes
- Hybridization probes: These typically single-stranded oligonucleotide probes used in hybridization-based detection techniques such as ISH and microarrays.
- PCR probes: Used for gene amplification or quantitative detection.
- Molecular beacons: Designed as hairpin structures, with a fluorescent dye and a quencher dye at opposite ends. The hairpin opens and emits fluorescence upon binding to complementary target sequences. These are commonly used for detecting and quantifying nucleic acid sequences.
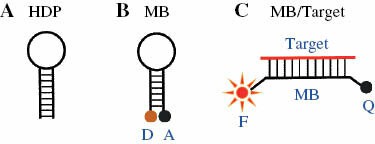 Fig. 1. Hairpin probes and basic detection mechanism (Huang J, Wu J, et al., 2015).
Fig. 1. Hairpin probes and basic detection mechanism (Huang J, Wu J, et al., 2015).
- Aptamers: Short single-stranded DNA sequences that fold into specific structures and bind specifically to proteins, small molecules, cells, or viruses, often used in disease diagnosis and therapy.
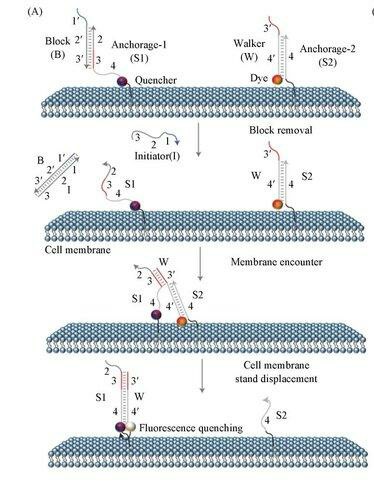 Fig. 2. Schematic illustration of using a DNA probe to monitor dynamic and transient molecular encounters on live cell membranes (Yang L, Miao Y, et al., 2020)
Fig. 2. Schematic illustration of using a DNA probe to monitor dynamic and transient molecular encounters on live cell membranes (Yang L, Miao Y, et al., 2020)
- Random probes: Also known as degenerate probes, they consist of combinations of random nucleotide sequences that may overlap with other target sequences. They are central to genomics, gene expression and molecular symbolism.
RNA probes
- Single-stranded cDNA probes: Probes derived via reverse transcription of mRNA, cloning and amplification. Applications include northern and southern blotting, ISH, microarray, diagnosis of diseases, cell sorting, and sequencing.
- cRNA probes: Internally generated complementary RNA sequences that resemble target sequences. Northern blotting, ISH, ribonuclease protection assay (RPA) and microarray analysis are typical applications.
- Oligonucleotide RNA probes: Artificially produced probes generated by DNA synthesizers, commonly used in northern blotting and ISH.
Applications in ISH (In Situ Hybridization)
DNA probes
DNA probes are frequently used in ISH technology to label and analyze specific genomic locations.
- Locus probes: Probes used to investigate individual genes or loci on the chromosome and copy number changes of target genes or loci.
- Alphoid probes: Telomere or centromere probes hybridise with highly repetitive sequences on telomeres and centromeres to detect numerical chromosomal abnormalities.
- Whole chromosome probes: Multi-color FISH probes are combinations of differentially labeled, long probes that can bind to different locations on different chromosomes. Mostly used in chromosomal rearrangement experiments.
- Microarray probes: Very short, stuck to rigid support surfaces, and widely used in high-throughput gene analysis.
RNA probes
RNA probes play a critical role in RNA in situ hybridization (RISH) and its derivative techniques, especially in studying gene expression and localizing specific mRNA molecules.
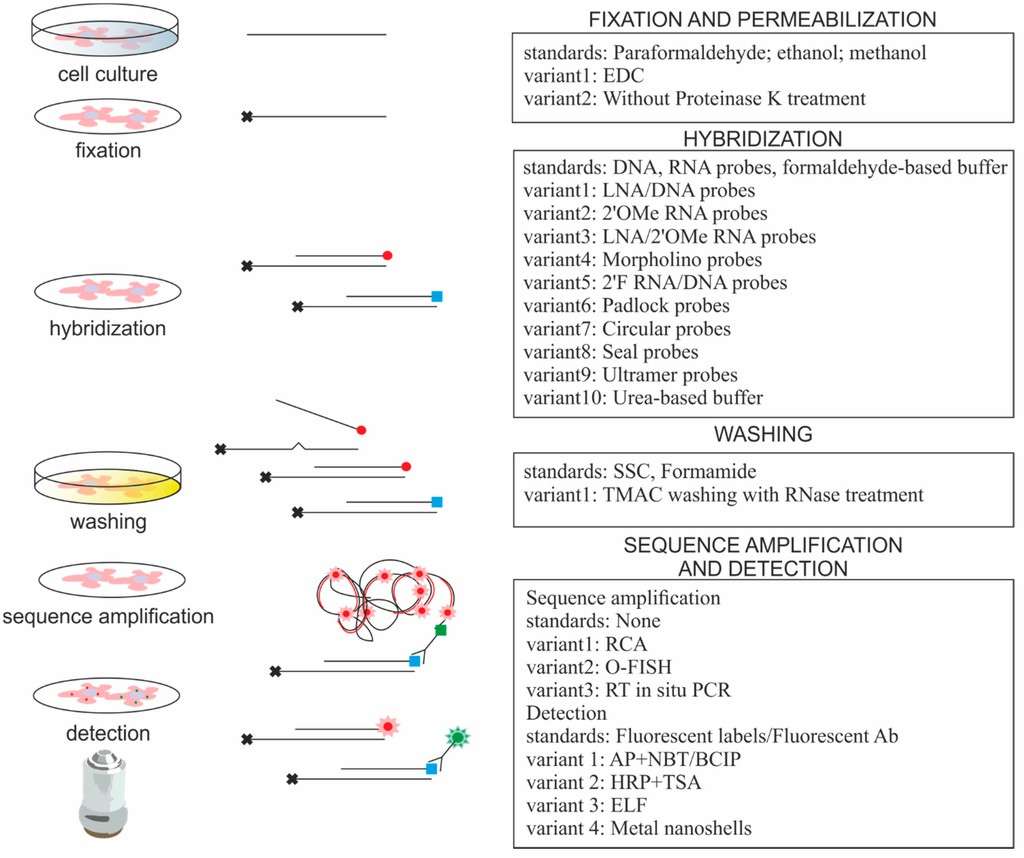 Fig. 3. In situ hybridization protocols used for imaging of small RNAs (Urbanek M O, Nawrocka A U, et al. 2015).
Fig. 3. In situ hybridization protocols used for imaging of small RNAs (Urbanek M O, Nawrocka A U, et al. 2015).
- Antisense RNA probes: Complementary to target mRNA, used to detect specific gene expression.
- Oligonucleotide probes: Primarily used to identify short RNA or RNA fragments during ISH.
- Specific cDNA probes: Capable of detecting DNA or RNA sequences with high stability and specificity but low hybridization efficiency, less used in ISH.
- Nested probes: Two separate single-stranded probes hybridize to the same target sequence from opposite directions, increasing the detection efficiency and signal strength, typically used with low-density RNA molecules.
- Locked nucleic acid (LNA) probes: Synthetic oligonucleotides with enhanced recognition power, typically used to detect expression and localization of miRNA and other short RNA molecules.
By thoroughly understanding the types and characteristics of DNA and RNA probes, researchers in molecular biology can better select and design experiments, thereby efficiently solving practical problems and advancing scientific discovery and technological innovation.
| Products & Services | Description |
| Fluorescent In Situ Hbridization (FISH) Service | Creative Bioarray offers a range of different FISH services including metaphase and interphase FISH, fibre-FISH, RNA-FISH, M-FISH, 3D-FISH, flow-FISH, FISH on paraffin sections, and immune-FISH. |
| FISH Probe Design, Synthesis, and Testing Service | Creative Bioarray is capable of developing custom FISH probes. Apart from that, we also offer mRNA ISH/FISH probes, miRNA ISH/FISH probes, and lncRNA ISH/FISH probes. |
References
- Huang J, Wu J, et al. Biosensing using hairpin DNA probes. Reviews in Analytical Chemistry, 2015, 34.
- Yang L, Miao Y, et al. DNA Nanotechnology on Live Cell Membranes. Chemical Research in Chinese Universities, 2020, 36(4).
- Urbanek M O, Nawrocka A U, et al. Small RNA Detection by in Situ Hybridization Methods. International Journal of Molecular Sciences, 2015, 16(6), 13259-13286.

