Fluorescence in situ hybridization (FISH) has become an indispensable tool in the life science industry, enabling the visualization and localization of specific nucleic acid sequences within intact cells and tissues. However, as with any advanced analytical method, mastering FISH can present its own set of challenges. That's why we've gathered our collective expertise to share a comprehensive guide on FISH tips and troubleshooting strategies, ensuring your research endeavors are met with consistent success.
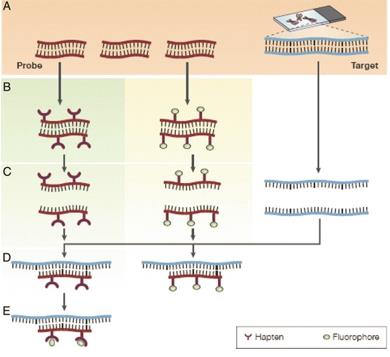 Fig. 1 The principles of fluorescence in situ hybridization. (Shakoori AR, 2017)
Fig. 1 The principles of fluorescence in situ hybridization. (Shakoori AR, 2017)
FISH Tips
When embarking on a FISH experiment, meticulous planning and execution are crucial for obtaining reliable and reproducible results. Here are some key tips to enhance the success of your FISH assays:
| Tips | Details |
| Sample preparation and fixation | - For cell samples, use healthy, actively growing cells to ensure good chromosome/nuclear morphology. For tissue samples, use fresh or minimally fixed material to preserve the target nucleic acid integrity.
- Choose an appropriate fixative, such as formaldehyde or paraformaldehyde, to preserve the cell morphology and maintain the integrity of the target nucleic acids.
- Ensure the fixation time and concentration are optimized to avoid over-fixation, which can reduce the accessibility of target sequences.
- For suspension cells, centrifuge and resuspend cells in the fixative. For adherent cells, fix cells directly on the slide or coverslip.
|
| Permeabilization | - Permeabilize the cells or tissues to allow the probe to access the target nucleic acids, using agents such as Triton X-100, Tween-20, or proteinase K.
- Optimize the permeabilization conditions to balance accessibility and preservation of cell morphology.
- Control permeabilization conditions such as concentration, time, and temperature to maintain sample integrity.
|
| Denaturation | - Denature the target nucleic acids (DNA or RNA) using heat or alkaline treatment to make them single-stranded and accessible for probe hybridization.
- Optimize the denaturation conditions to maintain the sample integrity and morphology.
|
| Probe preparation | - Design specific and sensitive FISH probes targeting the desired genomic regions or transcripts.
- Label the probes with fluorescent dyes (e.g., fluorescein, rhodamine, Cy3, Cy5) for detection.
- Optimize the probe concentration and hybridization conditions for efficient and specific binding.
|
| Probe application | - When applying FISH probes, try not to push down too hard on the coverslip as it can squeeze out of the sides, causing a patchy hybridization.
- Make sure the pipette tip is long enough so that the pipette itself doesn't come into contact when the tip reaches the bottom of the vial. Pipettes entering a probe vial may carry over probe reagent from one vial to the next resulting in cross contamination.
- When making FISH slides from fixed cell suspensions, use a template to ensure that the suspension is consistently spotted in the same place. Use the template for probe application also, which ensures the FlSH probe is applied in the correct place.
|
| Hybridization | - Incubate the prepared samples with the labeled FISH probes under appropriate hybridization conditions (temperature, time, and buffer composition).
- Use a humid chamber to prevent sample drying during hybridization.
|
| Post-hybridization washes | - Perform stringent washes to remove unbound or non-specifically bound probes.
- Adjust the wash conditions (temperature, salt concentration, and duration) to maintain the desired signal-to-noise ratio.
- Avoid prolonged wash times that may lead to probe detachment.
|
| Counterstaining | - Use a DNA-binding fluorescent dye (e.g., DAPI, propidium iodide) to counterstain the nuclei and visualize the overall cell or chromosome morphology.
- Counterstain after FISH hybridization and wash steps to avoid interference with probe binding.
|
Common Issues and Troubleshooting Strategies
Despite meticulous planning, FISH experiments may encounter various challenges that can impede accurate results. Here are common issues faced during FISH procedures and effective troubleshooting strategies to overcome them:
| Issues | Troubleshooting strategies |
| Poor or no signal | - Check the probe design and labeling efficiency.
- Optimize the denaturation and hybridization conditions.
- Increase the probe concentration or hybridization time.
- Ensure adequate permeabilization of the sample.
- Check the fluorescence microscope settings and filters.
|
| High background or non-specific signal | - Optimize the wash conditions (temperature, salt concentration, and duration).
- Increase the stringency of the washes.
- Ensure complete denaturation of the target DNA/RNA.
- Check for any cross-reactivity of the probe with non-target sequences.
- Optimize the probe concentration and hybridization time.
|
| Weak or faded signal | - Use a more sensitive fluorophore or consider signal amplification methods.
- Optimize the mounting medium and antifade reagents.
- Minimize exposure of the sample to light during imaging.
- Ensure the sample is not over-fixed or over-permeabilized.
|
| Uneven or patchy signal: | - Check for uniform distribution of the probe during hybridization.
- Ensure even permeabilization and denaturation of the sample.
- Avoid air bubbles or uneven drying during the mounting process.
- Verify the quality and consistency of the sample preparation.
|
| Morphological distortion or cell damage | - Optimize the fixation and permeabilization conditions.
- Avoid over-fixation or over-permeabilization.
- Use gentler methods for cell/tissue dissociation and spreading.
- Ensure appropriate handling and storage of the samples.
|
| Lack of reproducibility | - Standardize the sample preparation and FISH protocol steps.
- Use appropriate positive and negative controls.
- Ensure consistent reagent quality and storage conditions.
- Consider interlaboratory validation or proficiency testing.
|
| Unexpected or ambiguous results | - Verify the probe specificity and cross-reactivity.
- Check for potential sequence variations or chromosomal rearrangements.
- Correlate the FISH results with other techniques, such as qPCR or sequencing.
- Consult with experts or reference literature for guidance.
|
Creative Bioarray Relevant Recommendations
| Service/Product Types | Description |
| Fluorescent In Situ Hybridization (FISH) Service | Creative Bioarray offers a range of different FISH services including metaphase and interphase FISH (chromosomal assignment and clone ordering), Fibre-FISH (Chromosome Painting), RNA-FISH (cell-based gene expression assay), M-FISH (multicolor karyotyping), 3D-FISH (on three-dimensionally preserved nuclei), Flow-FISH (quantify the length of telomeres), FISH on paraffin sections (analysis of archive material), ImmunoFISH (combined FISH and IHC). |
| Probe | Creative Bioarray provides the most comprehensive list of FISH probes for rapid identification of a wide range of chromosomal aberrations across the genome. |
Reference
- Shakoori AR. (2017). "Fluorescence In Situ Hybridization (FISH) and Its Applications." Chromosome Structure and Aberrations. 343-67.
For research use only. Not for any other purpose.
 Fig. 1 The principles of fluorescence in situ hybridization. (Shakoori AR, 2017)
Fig. 1 The principles of fluorescence in situ hybridization. (Shakoori AR, 2017)
