FISH Techniques for Biofilm Detection
Biological membranes are complex structures composed of resident microorganisms, extracellular matrix, and the surrounding environment. These biofilms are prevalent in various fields, including healthcare, environmental science, and industry, which has spurred growing research interest in them. Modern molecular techniques such as next-generation sequencing (NGS) and RNA-seq offer advanced means to study the properties of biofilms. However, these techniques have a significant drawback: they disrupt the original spatial structure of the biofilm, making it impossible to visualize the specific locations of various components such as cells, genes, and metabolites within the biofilm. To address this challenge, fluorescence in situ hybridization (FISH) has emerged as one of the most widely used methods for studying the spatial distribution of biofilms.
Advantages of FISH for Biofilm Detection
FISH technology uses fluorescently labeled nucleic acid probes to hybridize with the rRNA, DNA, or mRNA of target microorganisms. Through fluorescence microscopy, the positions of these microorganisms within the biofilm can be directly observed. Compared to other molecular biology techniques, FISH offers multiple advantages:
- High spatial resolution: FISH can achieve precise spatial localization at the cellular, genetic, and metabolic levels, providing detailed images of the internal structure and species distribution within the biofilm.
- Non-destructive: FISH technology does not disrupt the 3D structure of the biofilm during detection, preserving the microorganisms' original physiological state. This is particularly important for studying microbial interactions.
- Quantitative analysis: When combined with digital image analysis, FISH can not only identify microbial species but also quantify their abundance, offering crucial data for functional studies of microbial communities within biofilms.
- Wide applicability: FISH can be used to detect samples from different environments, ranging from aquatic and soil biofilms in natural settings to pathogenic biofilms in clinical samples, providing accurate microbial distribution information.
Types of FISH Used for Biofilm Detection
- FISH: Classic FISH employs fluorescently labeled oligonucleotide probes that directly hybridize to the RNA or DNA of target microorganisms. This method is straightforward and flexible, making it suitable for basic microbial identification and localization. However, classic FISH may encounter issues of insufficient signal intensity and hindered probe diffusion in complex biofilm environments.
- CARD-FISH: Catalyzed reporter deposition (CARD)-FISH is an enhanced version of FISH that improves detection sensitivity through signal amplification. CARD-FISH uses fluorescent probes combined with horseradish peroxidase (HRP), significantly enhancing the detection signal. This is especially important for studying microorganisms in environmental samples like soil and sediment. However, CARD-FISH has poor permeabilization effects on cells, requiring optimized processing steps to ensure effective probe diffusion.
- NAM-FISH: Nucleic acid mimic (NAM)-FISH uses chemically modified DNA or RNA probes to enhance binding strength and specificity to the target sequence. Compared to traditional DNA probes, NAM probes like peptide nucleic acid (PNA) and locked nucleic acid (LNA) offer higher affinity and biological stability, providing stronger fluorescent signals in complex biofilm environments. Although NAM-FISH is complex and can distinguish a limited number of targets in a single experiment, its advantage lies in effective spatial characterization in thicker biofilms.
- FIVH: Fluorescence in vivo hybridization (FIVH) is a FISH variant designed specifically for in vivo biofilm research. Its advantage is that it does not require fixation and permeabilization steps, allowing detection in live tissues. This technology enables real-time observation of biofilm dynamics under different conditions without disrupting the biofilm structure. However, due to current equipment limitations, FIVH can only detect a few targets, with future improvements in imaging systems expected to enhance its multiplexing capabilities.
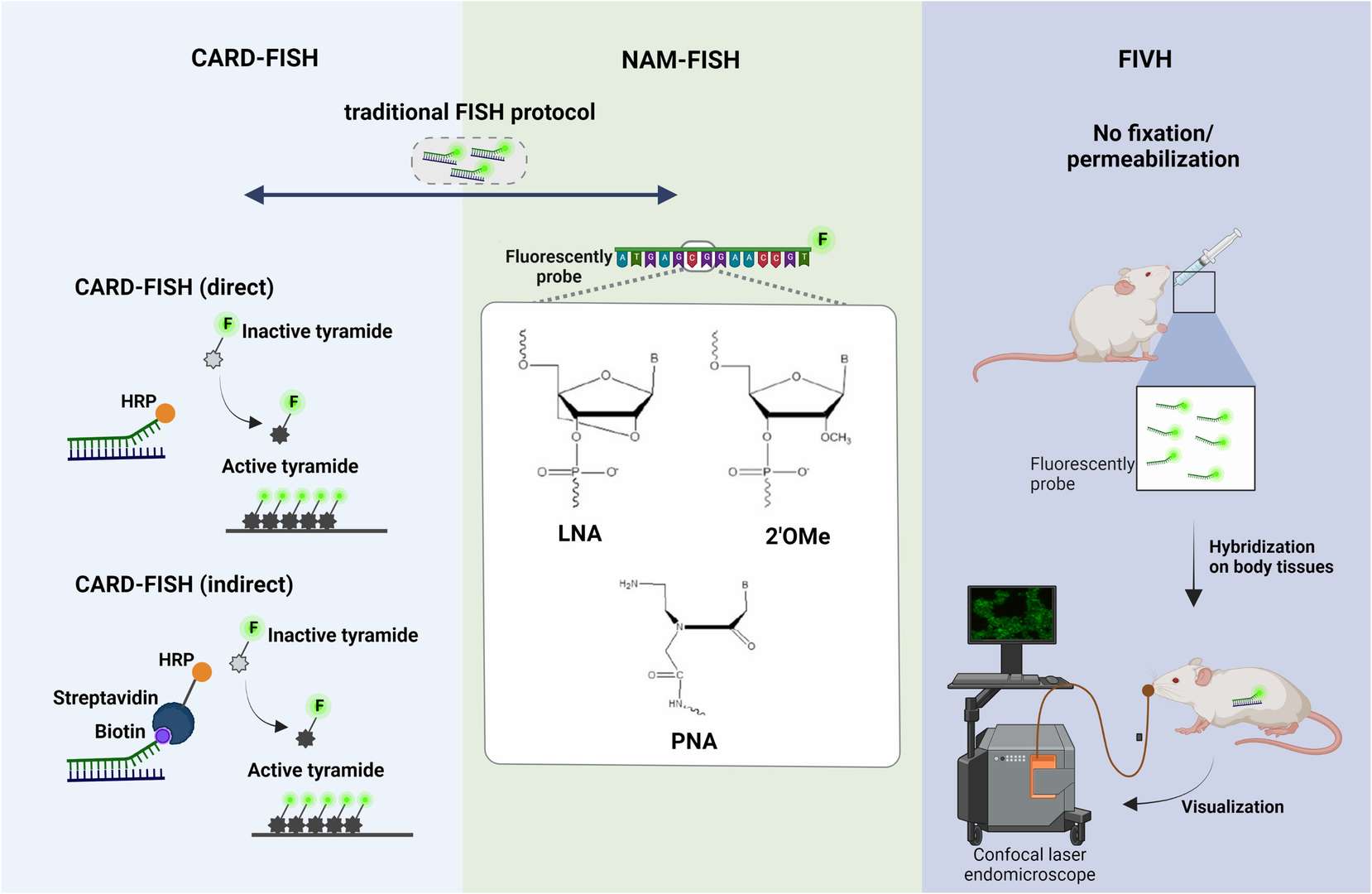 Fig. 1. Principle steps of (A) CARD-FISH, (B) NAM-FISH and (C) FIVH (Barbosa A, Miranda S, et al., 2023).
Fig. 1. Principle steps of (A) CARD-FISH, (B) NAM-FISH and (C) FIVH (Barbosa A, Miranda S, et al., 2023).
- DOPE-FISH: Double labeling of oligonucleotide probes (DOPE)-FISH is characterized by labeling both 5' and 3' ends of the probe with different fluorescent dyes, doubling the fluorescent signal compared to standard ISH. This method allows simultaneous detection of up to six different microbial species in a single experiment and significantly enhances signal strength without compromising detection specificity. DOPE-FISH shows strong potential for studying the structure and spatial distribution of multi-species biofilms.
- CLASI-FISH: Combinatorial labeling and spectral imaging (CLASI)-FISH is a technology capable of distinguishing and detecting numerous microbial species within a single biofilm sample. By using multiple fluorescent probe combinations and integrating spectral imaging, CLASI-FISH achieves multiplexed detection and spatial analysis of complex biofilm communities. Despite the complexity of probe design, this technology holds great potential for studying microbial ecosystems.
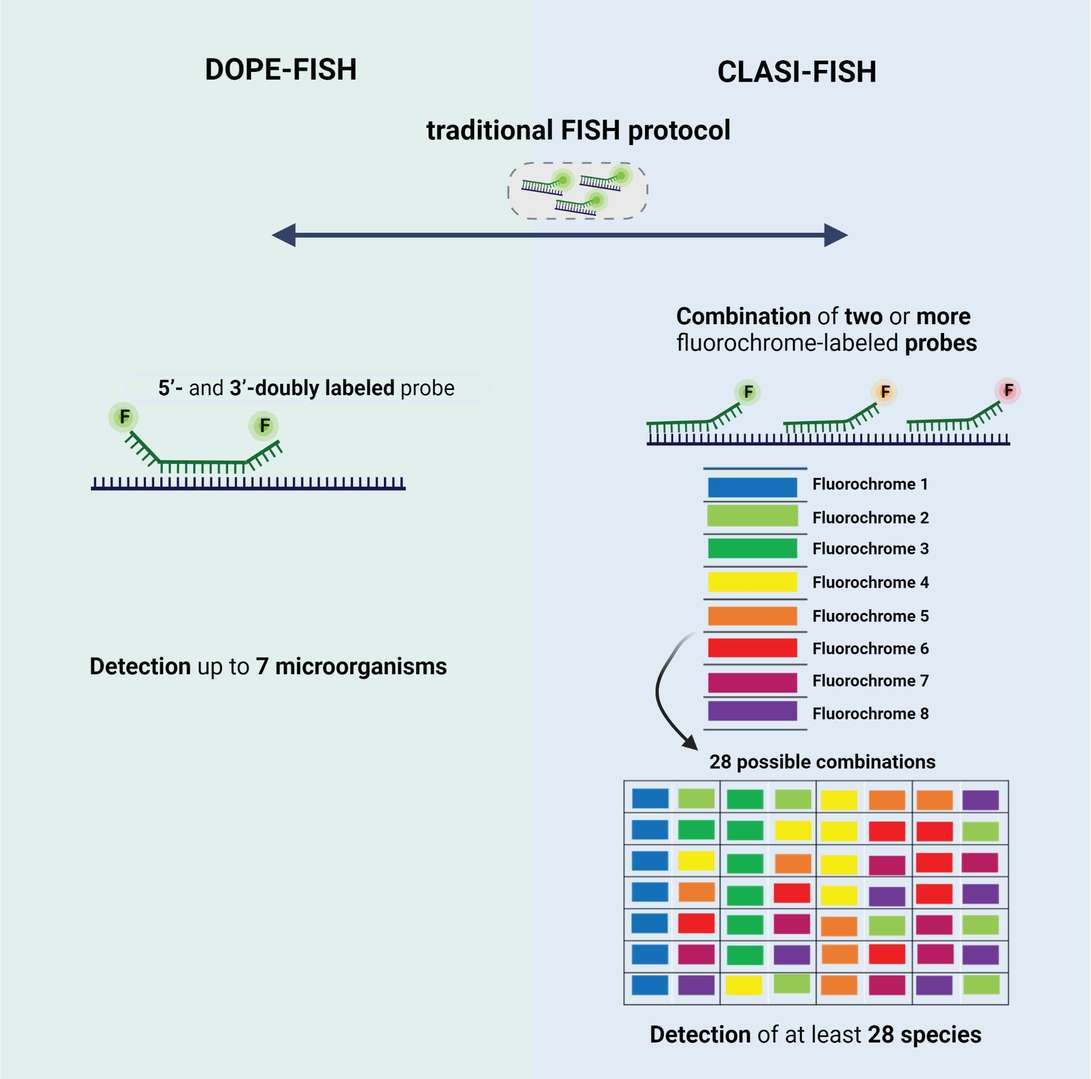 Fig. 2. Schematic representation of type of probes used in (A) DOPE-FISH and (B) CLASI-FISH. (Barbosa A, Miranda S, et al., 2023).
Fig. 2. Schematic representation of type of probes used in (A) DOPE-FISH and (B) CLASI-FISH. (Barbosa A, Miranda S, et al., 2023).
- HiPR-FISH : Highly multiplexed spatial mapping of microbial communities via hybridization chain reaction (HiPR)-FISH employs a two-step hybridization method and automated image segmentation to achieve higher classification resolution and multiplexing. This technique uses a combination of coding probes and readout probes, greatly increasing the number of detectable microbial species. HiPR-FISH allows precise detection and spatial analysis of multiple targets in a single experiment, making it highly suitable for studying complex microbial biofilm communities.
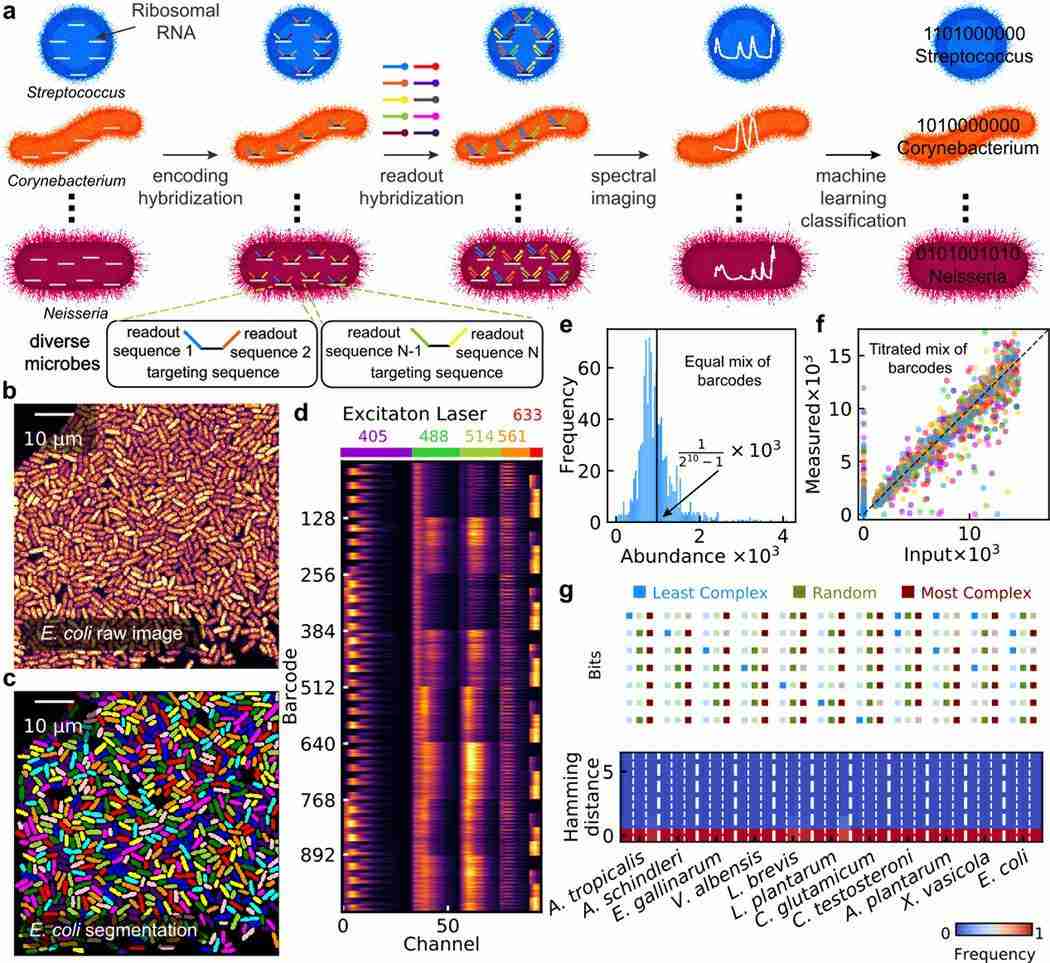 Fig. 3. HiPR-FISH working principle (Shi H, Shi Q, et al., 2020).
Fig. 3. HiPR-FISH working principle (Shi H, Shi Q, et al., 2020).
- MAR-FISH: Microautoradiography (MAR)-FISH combines FISH technology with radioactive isotope labeling to analyze microbial metabolic activity at the single-cell level. By detecting the assimilation of specifically labeled substrates, MAR-FISH can identify and analyze active microorganisms within biofilms under in situ conditions. However, this technology involves radiation risks and complex sample preparation.
- FISH-NanoSIMS: This technique integrates FISH with nanoscale secondary ion mass spectrometry, addressing the limitations of MAR-FISH regarding radioactive element types and quantitative accuracy. NanoSIMS analyzes sample elements at high resolution and, combined with FISH, enables simultaneous detection of microbial identity and metabolic activity. This technique offers significant advantages in studying environmental microbial communities, although it requires expensive equipment and complex sample preparation.
- BONCAT-FISH: Bio-orthogonal noncanonical amino acid tagging (BONCAT)-FISH achieves analysis of microbial translation activity by incorporating non-canonical amino acids in vivo. This method does not require isotope labeling and uses click chemistry to fluorescently label proteins containing non-canonical amino acids, combining with FISH for in situ detection. Although costly and time-consuming, this method has important applications in studying dynamic changes in complex microbial communities.
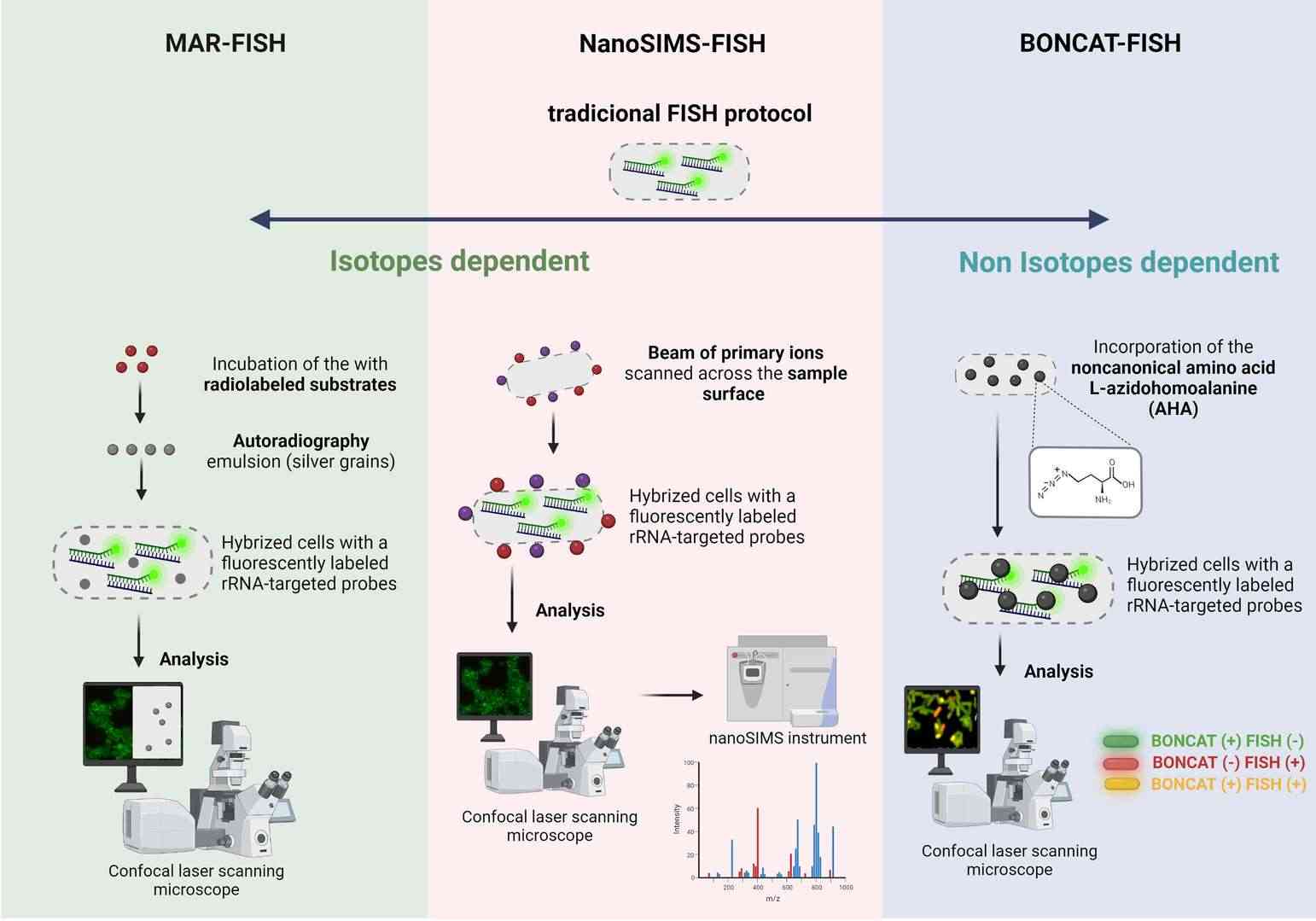 Fig. 4. Schematic representation of (A) MAR-FISH, (B) NanoSims-FISH and (C) BONCAT-FISH (Barbosa A, Miranda S, et al., 2023).
Fig. 4. Schematic representation of (A) MAR-FISH, (B) NanoSims-FISH and (C) BONCAT-FISH (Barbosa A, Miranda S, et al., 2023).
Conclusion
Fluorescence in situ hybridization (FISH) technology demonstrates strong application potential in biofilm research. Through various FISH variants, researchers can analyze the spatial distribution of microorganisms in situ, revealing the metabolic activities and species diversity within biofilms. With continuous technological advancements, the application of FISH in healthcare, environmental science, and industry will become increasingly widespread, providing scientists with more precise and comprehensive research tools.
| Products & Services | Description |
| FluorescentIn Situ Hbridization (FISH) Service | Creative Bioarray offers a range of different FISH services including metaphase and interphase FISH, fibre-FISH, RNA-FISH, M-FISH, 3D-FISH, flow-FISH, FISH on paraffin sections, and immune-FISH. |
| FISH Analysis of Microorganisms | Creative Bioarray is one of the well-recognized experts who is professional in FISH technologies for a broad range of project objective. With years of experience, our scientists can offer high-quality FISH analysis of microorganisms to meet your demands. |
| CARD-FISH for Environmental Microorganisms | With many years of experience and in-depth knowledge, Creative Bioarray can offer the CARD-FISH service with different kinds of samples. |
References
- Barbosa, A., et al. Imaging biofilms using fluorescence in situ hybridization: seeing is believing. Frontiers in cellular and infection microbiology, 2023. 13, 1195803.
- Shi H, et al. Highly multiplexed spatial mapping of microbial communities. Nature. 2020 Dec;588(7839):676-681.
- Frickmann, H., et al. Fluorescence in situ hybridization (FISH) in the microbiological diagnostic routine laboratory: a review. Critical reviews in microbiology, 43(3), 263–293.