Guidelines for the Design of FISH Probes
Fluorescence in situ hybridization (FISH) is a well-established technique that allows the detection of microorganisms in diverse types of samples (e.g., clinical, food, environmental samples, and biofilm communities). The FISH probe design is an essential step in this technique. For this, two strategies can be used, the manual form based on multiple sequence alignment to identify conserved regions and programs/software specifically developed for the selection of the sequence of the probe. Additionally, databases/software for the theoretical evaluation of the probes in terms of specificity, sensitivity, and thermodynamic parameters (melting temperature and Gibbs free energy change) are used.
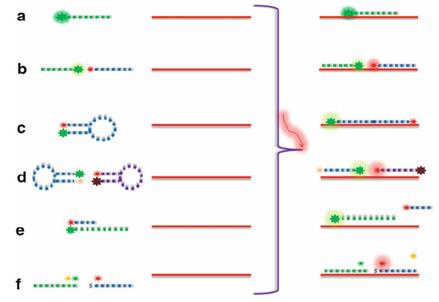 Fig.1 Approaches to the design of fluorescent probes for FISH experiments. (Kadam US, et al., 2013)
Fig.1 Approaches to the design of fluorescent probes for FISH experiments. (Kadam US, et al., 2013)
Probe Design Considerations
The probe design/selection is one of the most important factors in the successful performance of FISH. The selection of the optimal probe must take into account several parameters:
Probe length
Since the specificity, hybridization temperature, and time are partly dependent on probe length, this parameter is critical for successful hybridization. Probes typically have 15-30 nucleotides in length. If the probe is too long, the selectivity toward the target decreases, and the possibility of the formation of intramolecular structures increases; in addition, low synthesis yields, longer hybridization times, and the cost of the synthesis of the probe is greater. Conversely, probes with few bp in length might lack specificity; the target sequence might be present in a larger number of biological targets. The length of DNA probes is at least 18 nucleotides, PNA probes can contain 13-18 bases, and LNA probes can contain 10-15 nucleotides in length.
Overall Gibbs free energy change (ΔG0 overall)
ΔG0 overall is a thermodynamic parameter that can be used as an indicator of whether a reaction is thermodynamically favorable or not. A negative ΔG0 value (ΔG0 < 0) is an indicator of a thermodynamically favorable reaction; the more the negative value, the more the favorable reaction. It is well-established that for DNA probes ΔG0 should be between -13 and -20 kcal/mol to maximize hybridization efficiency without compromising specificity.
Melting temperature (Tm)
Tm is the temperature at which 50% of the nucleic acid strands form a double helix and the other 50% are single-stranded. A higher predicted Tm usually provides better results than probes with lower Tm.
Pyrimidine content (GC)
The GC content might provide information about the strength of hybridization. For probes with a low GC content, it may be necessary to extend the probe sequence to keep the Tm in the recommended range since GC content and Tm are strictly dependent on one another.
Complementary probe sequences
Self-complementary regions with more than three nucleotides within the probe should be avoided.
Secondary structure
The presence of secondary structures is an important factor when designing a probe. This greatly decreases the number of nucleotides available for binding in the reaction. The presence of hairpin loops in the probe reduces the FISH efficiency, limiting the probe's ability to bind to the target site.
Specificity
A high specificity value means that the probe only detects the target for which it was designed.
Sensitivity
A high sensitivity value means that the probe detects all strains of the target taxa for which the probe was designed.
Probe Design Algorithms and Software Tools
| Software tools | Advantages |
| Primer3 |
|
| OligoCalc |
|
| ArrayDesign |
|
Challenges and Troubleshooting
Handling GC-rich or AT-rich Target Sequences
Target sequences with extreme GC or AT content can pose significant design challenges, as they can impact probe stability, specificity, and hybridization efficiency.
- Comprehensive sequence analysis. Our advanced computational tools thoroughly evaluate the target region, identifying potential problematic areas and optimizing probe sequences accordingly.
- Thermodynamic modeling. We leverage sophisticated thermodynamic models to predict the behavior of probes under various hybridization conditions, allowing us to fine-tune design parameters.
- Empirical optimization. When necessary, we conduct experimental validation and iteration to refine probe design, ensuring robust performance even in the face of challenging target sequences.
Addressing Issues with Probe Accessibility and Hybridization
Effective FISH probe design requires considering the complex three-dimensional structure of target DNA or RNA, as well as potential interference from secondary structures or DNA-binding proteins.
- In silico secondary structure prediction. Our computational tools analyze target sequences to identify and avoid regions with stable secondary structures that may impede probe accessibility.
- Probe design optimization. We carefully select probe lengths, positions, and labeling strategies to maximize target accessibility and hybridization efficiency.
Strategies for Improving Signal Intensity and Resolution
Obtaining high-quality FISH signals with clear cellular localization and resolution is crucial for accurate data interpretation.
- Probe labeling and design. We explore various labeling strategies, such as direct versus indirect labeling, to optimize signal intensity and specificity.
- Image acquisition and processing. Our team collaborates closely with clients to ensure optimal microscopy settings and image analysis workflows to maximize the quality of FISH data.
- Experimental optimization. We work with clients to fine-tune sample preparation, hybridization conditions, and other experimental parameters to further improve FISH signal resolution and intensity.
Creative Bioarray Relevant Recommendations
| Service/Product Types | Description |
| FISH Probe Design, Synthesis, and Testing Service | With access to the entire RP11 BAC library, Creative Bioarray is capable of developing custom FISH probes. Apart from that, we also offer mRNA ISH/FISH Probes, miRNA ISH/FISH Probes, and lncRNA ISH/FISH Probes. |
| Probe | Creative Bioarray provides the most comprehensive list of FISH probes for rapid identification of a wide range of chromosomal aberrations across the genome. |
Reference
- Kadam US, et al. (2013). "Gene expression analysis using conventional and imaging methods." DNA and RNA Nanobiotechnologies in Medicine: Diagnosis and Treatment of Diseases. 141-162.

