Featured Products
Our Promise to You
Guaranteed product quality, expert customer support

ONLINE INQUIRY
- Specification
- Background
- Scientific Data
- Q & A
- Customer Review
The SKN-3 cell line is a human oral squamous carcinoma cell line that was established through xenotransplantation in mice. This method involves implanting human cancer cells into a mouse model, which provides a suitable environment for the cells to grow and proliferate. SKN-3 originated from a patient with oral squamous cell carcinoma, a common type of head and neck cancer known for its aggressive nature and propensity for metastasis.
One of the advantages of the SKN-3 cell line is its ability to closely mimic the in vivo environment of oral squamous carcinoma. This makes it particularly valuable for preclinical studies, allowing researchers to evaluate tumor behavior, interactions with the immune system, and the effectiveness of various therapeutic agents.
In addition to its application in drug testing, SKN-3 has been instrumental in investigating the molecular mechanisms underlying oral squamous cell carcinoma. Researchers have employed this cell line to explore cellular signaling pathways, gene expression profiles, and the role of various factors in tumor development and metastasis. The findings from studies using SKN-3 contribute to our understanding of the disease, potentially leading to more effective diagnostic markers and therapeutic strategies.
Detailed Mapping of the 19q13.12-13.2 Amplicon in SKN-3 Cells
Using a high-resolution array-CGH with an in-house BAC array, the region 19q13.12-13.2 is to be amplified in one of 18 OSCC cell lines, SKN-3. To map this region in detail, FISH was performed using 14 sequential BAC clones (RP11-44B13, 115H16, 587I9, 91H20, 140E1, 118P21, 446K10, 787F3, 256O9, 841P21, 343J4, 67A5, 246P10, and 384E6; Fig. 1a) as probes distributing across approximately 3.5 Mb and found remarkable amplification with a homogenously staining region (HSR), on the metaphase chromosomes of SKN-3 cells (Fig. 1a, b). This amplicon is bounded by two BACs, RP11-587I9 and RP11-67A5, whereas three BACs, namely RP11-44B13, RP11-115H16, and RP11-384E6, showed lower copy-number gain comparatively (Fig. 1b), helping us to map the amplicon to a 2.5‐Mb region (Fig. 1a). Since the oncogene AKT2, a putative and well‐characterized target for amplification within 19q13.12‐13.2 in other tumors, is located at the edge of this amplicon in SKN-3 cells (Fig. 1a, b), whether AKT2 is included in this amplicon as a potential driver gene was examined. High-density oligonucleotide array-CGH and quantitative genomic PCR analyses demonstrated that most but not the entire AKT2 gene was included within the amplicon (Fig. 1c, d). This result was confirmed by the relatively low level of AKT2 mRNA in SKN-3 cells compared with other OSCC cell lines without the 19q13.12-13.2 amplicon. Conversely, the defined 19q13.12-13.2 amplicon in SKN-3 contains 66 transcripts, including 49 known genes and 17 possible transcripts encoding hypothetical or predicted proteins, between ZNF573 and CNTD2 based on the human genome databases, prompting us to explore novel targets for amplification other than the AKT2 gene.
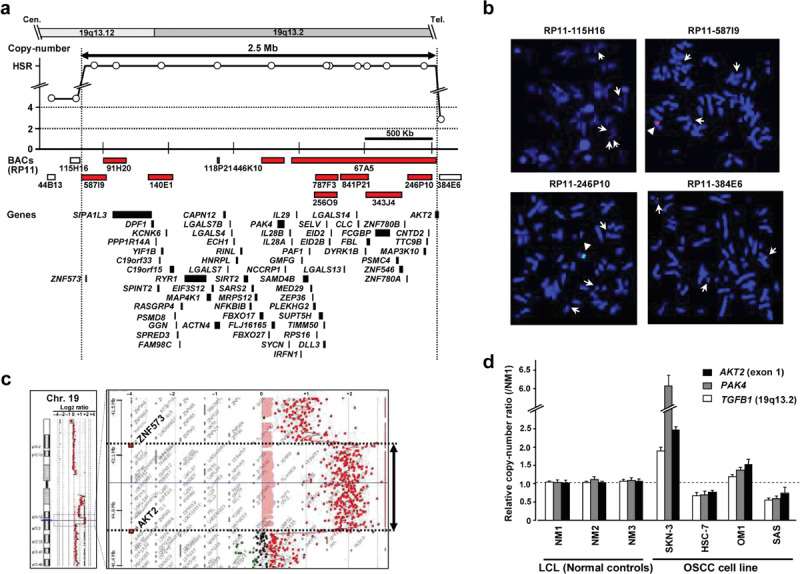 Fig. 1 Genetic aberrations at 19q13.12-13.2 in the oral squamous cell carcinoma (OSCC) cell line SKN-3. (a) Amplicon map of genomic profiles of 19q13.12-13.2 amplification in the SKN-3 cell line. (b) Representative images of the FISH analysis of metaphase chromosomes prepared from SKN-3 cells. (c) Changes in copy number around 19q13.12-13.2 in SKN-3 cells were determined using high-density oligonucleotide array-CGH. (d) Representative results of quantitative real-time genomic PCR for the AKT2 (exon1), PAK4, and control TGFB1 (19q13.2) genes. (Begum A, et al., 2009)
Fig. 1 Genetic aberrations at 19q13.12-13.2 in the oral squamous cell carcinoma (OSCC) cell line SKN-3. (a) Amplicon map of genomic profiles of 19q13.12-13.2 amplification in the SKN-3 cell line. (b) Representative images of the FISH analysis of metaphase chromosomes prepared from SKN-3 cells. (c) Changes in copy number around 19q13.12-13.2 in SKN-3 cells were determined using high-density oligonucleotide array-CGH. (d) Representative results of quantitative real-time genomic PCR for the AKT2 (exon1), PAK4, and control TGFB1 (19q13.2) genes. (Begum A, et al., 2009)
Normal squamous cells become cancerous when their DNA changes and cells begin growing and multiplying. Over time, these cancerous cells can spread to other areas inside of your mouth and then to other areas of your head and neck or other areas of your body.
Ask a Question
Average Rating: 4.0 | 1 Scientist has reviewed this product
Long-term storage
The tumor cell products offer long-term storage options, allowing us to maintain a consistent supply of high-quality tumor cells for extended periods.
27 June 2023
Ease of use
After sales services
Value for money
Write your own review
- You May Also Need