Featured Products
Our Promise to You
Guaranteed product quality, expert customer support

Esophageal Tumor Cells
- Product List
- Background
- Applications
- Scientific Data
- FAQ
Esophageal cancer, including tumor cells in the esophagus, is a complex and multifaceted disease, encompassing different histologic types such as squamous cell carcinoma and adenocarcinoma. Esophageal tumor cells, often associated with esophageal cancer, represent a critical area of research within the domain of oncology. The human esophagus, a muscular tube that connects the throat to the stomach, can be afflicted by the uncontrolled growth and division of abnormal cells, resulting in the formation of tumors. These cells may exhibit various pathological features, leading to significant morbidity and mortality in affected individuals. The understanding and characterization of esophageal tumor cells are vital for developing diagnostic, therapeutic, and preventive strategies to combat esophageal cancer.
Characteristics of Esophageal Tumor Cells
- Cell morphology and structure. Microscopic examination of esophageal tumor cells has shown that they often display an irregular, pleomorphic shape with a larger nucleus-to-cytoplasm ratio compared to normal esophageal epithelial cells. Additionally, an increased number of nucleoli within the tumor cell nuclei is indicative of their high transcriptional activity and protein synthesis demands. Interestingly, esophageal tumor cells also demonstrate alterations in their cytoskeletal organization, with disrupted actin filaments and microtubule networks. These structural changes contribute to the enhanced motility and invasive capabilities of the tumor cells, allowing them to break through the basement membrane and infiltrate surrounding tissues.
- Proliferation and growth patterns. One of the hallmarks of esophageal tumor cells is their uncontrolled proliferation and aggressive growth patterns. These cells possess a significantly higher proliferation rate compared to normal esophageal cells, driven by dysregulation in key cell cycle regulatory pathways. Furthermore, esophageal tumor cells exhibit a remarkable ability to adapt to their microenvironment, thriving even in nutrient-deprived or hypoxic conditions.
- Genetic and molecular alterations. Recurrent mutations are discovered in key genes involved in cell cycle regulation, such as TP53, CDKN2A, and KRAS. These genetic alterations lead to the disruption of normal cell cycle checkpoints, enabling uncontrolled cell division and evasion of apoptosis. Additionally, epigenetic modifications, including aberrant DNA methylation patterns and histone modifications, can silence the expression of tumor suppressor genes and promote the activation of oncogenic pathways. Esophageal tumor cells often exhibit hyperactivation of pathways like the PI3K/Akt/mTOR axis and the MAPK/ERK pathway, which contribute to their aggressive phenotype and metastatic potential.
Research and drug development
Esophageal tumor cells serve as invaluable tools for preclinical research and drug development. These In vitro models utilizing tumor cells facilitate the evaluation of potential therapeutic agents, elucidation of drug resistance mechanisms, and discovery of novel molecular targets.
Biomarker discovery
The analysis of esophageal tumor cells contributes to the identification and validation of biomarkers with diagnostic, prognostic, and predictive utility. Biomarker discovery within tumor cells aids in the development of non-invasive diagnostic assays, refinement of patient stratification algorithms, and monitoring of treatment response.
Therapeutic implications
The study of esophageal tumor cells is indispensable for the development of novel therapeutic modalities. Targeted therapies, immunotherapies, and precision medicine approaches are being explored to combat esophageal cancer, with a focus on eradicating tumor cells while minimizing adverse effects on normal tissues. Understanding the molecular aberrations and signaling pathways within esophageal tumor cells has the potential to revolutionize treatment strategies and improve patient outcomes.
Common Expression Programs of Epithelial Cells in ESCC Tumors
Esophageal squamous-cell carcinoma (ESCC), one of the most prevalent and lethal malignant diseases, has a complex but unknown tumor ecosystem. The composition of ESCC tumors was investigated based on 208,659 single-cell transcriptomes derived from 60 individuals. 8 expression programs were identified with different functions and cell status (Fig. 1a, b) and defined cells expressing ≥70% of genes in a given program as program cells.
The most activated pathways in the program cells were selected by comparison with non-program cells to analyze their functions (Fig. 1c). The mucosal immunity-like (Mucosal) program was characterized by the expression of genes associated with innate immune response (e.g., S100P) and mucosal defensive mechanisms including mucosal chemokine (e.g., CXCL17) and mucus production (e.g., AGR2 and MUC20). The stress responses (Stress) program consisted of immediate early genes (e.g., EGR1, JUN, and FOS) that are activated in response to widespread cellular stimuli and displayed upregulation of TNFα signaling, UV response, p53, and apoptosis pathways. The antigen presentation (AP) program had increased expression of major histocompatibility complex (MHC) class II molecules (e.g., CD74, HLA-DPA1, and HLA-DRA/B1/B5) that are involved in initiating adaptive antitumor immune responses. The cell cycle (Cycling) program was characterized by high expression of genes involved in cell proliferation (e.g., CENPW, CKS1B, and BIRC5) and presented activation of the E2F targets, G2M checkpoint, and MYC targets pathways, suggesting tumor cell proliferation. Two programs were associated with epithelial differentiation (Epi1/2). The Epi1 program was characterized by the expression of stress keratins (KRT6, KRT16, and KRT17) that are associated with keratinocyte hyperproliferation and therefore may play a role in enhancing tumorigenesis and tumor growth. The Epi2 program had the overexpressed genes related to the terminal differentiation such as envelope proteins (SPRR1A/1B) and calprotectin (S100A8/9), apical surface, the PI3K/AKT/mTOR signaling, the complement, and p53 pathways. The mesenchymal cell-like properties (Mes) program consisted of genes such as VIM and SPARC and showed activation of epithelial-mesenchymal transition (EMT) and angiogenesis pathways. Finally, the oxidative stress or detoxification (Oxd) program was characterized by the expression of multiple peroxidases and reductases (e.g., GPX2 and AKR1C1) involved in the defense against oxidative damage.
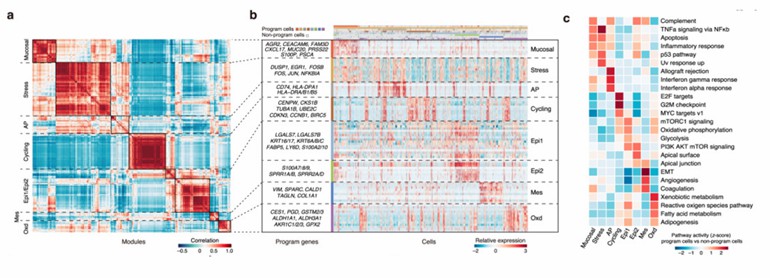 Fig. 1 Identification of eight common expression programs of epithelial cells in ESCC tumors. (Zhang X, et al, 2021)
Fig. 1 Identification of eight common expression programs of epithelial cells in ESCC tumors. (Zhang X, et al, 2021)
Expression of CD44 in ESCC Specimens
ESCC is a major subtype of esophageal cancer causing significant morbidity and mortality. The mechanism of initiation and progression of this disease is unclear. To investigate whether the potential ESCC TICs with high expression levels of CD44 can be detected in clinical specimens, CD44 expression was examined in 171 paraffin-embedded ESCC tissues by IHC staining.
In normal esophageal tissues, CD44 was found to be heterogeneously expressed. Among all the ESCC specimens, 30% (52/171) were either negative or weakly positive for CD44, and 70% (119/171) were CD44 strongly positive. The strongest expression of CD44 was found in the basal layer where normal esophageal stem cells are localized (Fig. 2A, B). Although no association was found between CD44 expression and clinicopathological parameters of ESCC patients, except for the degree of cellular differentiation, heterogeneous expression of CD44 was noted in most specimens. Keratin pearls are structures that occur during terminal differentiation of ESCC cells; these cells were either weakly positive or negative for CD44 expression (Fig. 2C). For most samples, most of the cells were uniformly strong expressers of CD44; however, a minority of cells within these samples had low expression of CD44 (Fig. 2C, D). A similar heterogeneity of expression occurred in specimens with low average weak expression of CD44; nevertheless, some of the cells strongly expressed CD44 (Fig. 2E, F). These results suggest that potential ESCC TICs with high expression levels of CD44 may exist in clinical ESCC samples.
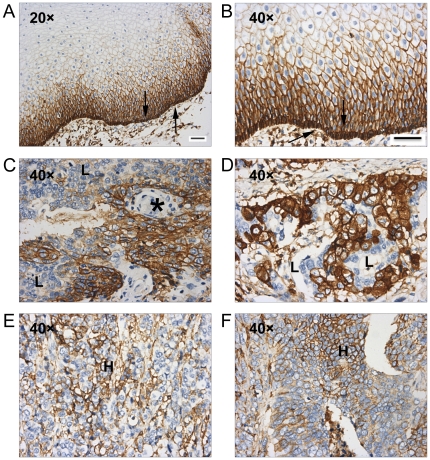 Fig. 2 Representative IHC analyses of CD44 in ESCC specimens. (Zhao JS, et al., 2011)
Fig. 2 Representative IHC analyses of CD44 in ESCC specimens. (Zhao JS, et al., 2011)
All of our cell lines are validated by STR.
The tumor microenvironment plays a crucial role in shaping the behavior of esophageal tumor cells. The interplay between the tumor cells and the surrounding stromal components, such as fibroblasts, immune cells, and the extracellular matrix, can promote tumor cell proliferation, invasion, and metastasis, suppress the immune system's anti-tumor response, provide a supportive niche for tumor cell survival and growth.
Esophageal tumor cells often harbor various genetic and molecular alterations, including mutations in key cell cycle regulatory genes, such as TP53, CDKN2A, and KRAS; epigenetic modifications, including aberrant DNA methylation and histone modifications; dysregulation of signaling pathways, like the PI3K/Akt/mTOR and MAPK/ERK cascades.
Description: moderate differenciated squamous cell carcinoma, stage 3
Description: Human squamous cell carcinoma cell line established from recurrent cancer (esophageal cancer).
Description: Human moderately differentiated squamous cell carcinoma cell line established from esophageal cancer.
Description: Human moderately differentiated squamous cell carcinoma cell line established from esophageal cancer.
Description: Human moderately differentiated squamous cell carcinoma cell line established from esophageal cancer.
Description: Human squamous cell carcinoma cell line established from esophageal cancer.
Description: Human moderately differentiated squamous cell carcinoma cell line established from esophageal cancer.
Description: Human moderately differentiated squamous cell carcinoma cell line established from esophageal cancer.
Description: Human moderately differentiated squamous cell carcinoma cell line established from esophageal cancer.
Description: Human moderately differentiated squamous cell carcinoma cell line established from esophageal cancer.
Description: Human moderately differentiated squamous cell carcinoma cell line established from esophageal cancer.
Description: Human moderately differentiated squamous cell carcinoma cell line established from esophageal cancer.
Description: Human poorly differentiated squamous cell carcinoma cell line established from esophageal cancer.
Description: Human poorly differentiated squamous cell carcinoma cell line established from esophageal cancer.
Description: Human poorly differentiated squamous cell carcinoma cell line established from esophageal cancer.
Description: Human moderately differentiated squamous cell carcinoma cell line established from esophageal cancer.
Description: Human squamous cell carcinoma from esophagus cancer.
Description: Human moderately differentiated squamous cell carcinoma cell line established from esophageal cancer.

