Featured Products
Our Promise to You
Guaranteed product quality, expert customer support

ONLINE INQUIRY
COLO-800
Cat.No.: CSC-C0319
Species: Human
Source: melanoma
Morphology: epithelial-like cells growing as monolayer; some cells look like fibroblasts or neurons
Culture Properties: monolayer
- Specification
- Background
- Scientific Data
- Q & A
- Customer Review
Immunology: cytokeratin -, desmin -, endothel -, GFAP -, HMB-45 +, neurofilament -, vimentin +
Viruses: ELISA: reverse transcriptase negative; PCR: EBV -, HBV -, HCV -, HHV-8
The COLO-800 cell line, established in the late 1970s from a metastatic malignant melanoma of a female patient, serves as a critical model for studying NRAS-driven melanomas. Characterized by epithelial-like morphology and adherent growth, COLO-800 harbors an oncogenic NRAS Q61K mutation, distinguishing it from BRAF-mutant melanomas and making it particularly relevant for researching this genetically distinct subset, which constitutes 15-20% of cases. Current research leverages these characteristics to explore targeted therapies against the MAPK pathway, such as MEK inhibitors, and combinatorial approaches with CDK4/6 inhibitors or immunotherapies (e.g., anti-PD-1/PD-L1 agents), addressing the limited efficacy of BRAF inhibitors in NRAS-mutant contexts. Additionally, COLO-800's adherent nature facilitates high-throughput drug screening, invasion assays, and 3D spheroid models to study metastasis and microenvironment interactions. Recent studies also employ this cell line to dissect resistance mechanisms and evaluate novel therapeutic synergies in xenograft models, underscoring its utility in advancing precision oncology for aggressive, treatment-resistant melanomas.
Melanoma Cancer Cell Lines: A Model of Cancer Metabolic Heterogeneity
To investigate the different types of tumor metabolic rewiring in melanoma, three human melanoma cell lines are analyzed: COLO 800 (derived from a primary subcutaneous nodule), COLO 794 (derived from a subcutaneous metastasis from the same individual as COLO 800), and A375 (derived from a metastatic nodule from a different individual). Consistent with their shared origin, COLO 800 and COLO 794 exhibited similar proliferation rates and metabolic behaviors in contrast to A375 (Fig. 1A, B). Despite all three cell lines displaying comparable glucose dependency (Fig. 1D), the untargeted metabolic profiling revealed different glucose utilization rates (Fig. 1E). Moreover, the increased levels of amino acids, such as arginine, proline, cysteine, asparagine, lysine, and methionine, in COLO 800 and COLO 794 compared with A375 further emphasize their reliance on distinct metabolic programs to maintain anabolic processes (Fig. 1E). On the other hand, the A375 cells exhibited an enrichment of ketone bodies, a well-known metabolic adaptation to cellular stress or nutrient deprivation (Fig. 1F). Furthermore, the elevated basal ROS levels (Fig. 1G) and reduction in redox scavenger metabolites observed in the A375 cells may contribute to their enhanced proliferative capacity (Fig. 1A).
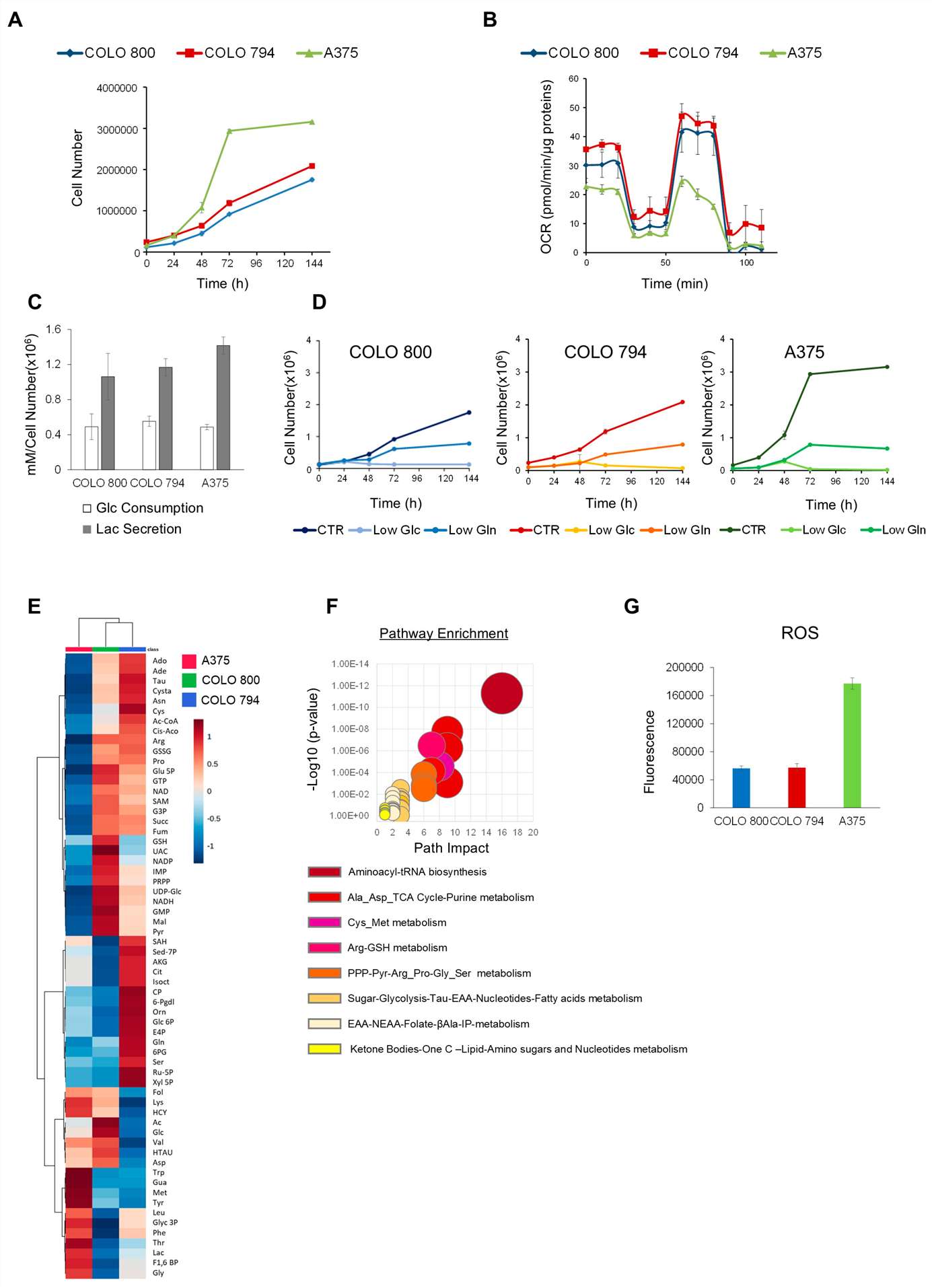 Fig. 1. Metabolic phenotype characterization of melanoma cell lines (Baldassari, Federica, et al. 2025).
Fig. 1. Metabolic phenotype characterization of melanoma cell lines (Baldassari, Federica, et al. 2025).
CXCR4 is Associated with RUNX2 Expression, Melanoma Invasiveness, and Osteotropism
Overexpression of the Runt-related transcription factor 2 (RUNX2) has been reported in several cancer types, and the C-X-C motif chemokine receptor 4 (CXCR4) has an important role in tumour progression. However, the interplay between CXCR4 and RUNX2 in melanoma cells remains poorly understood. In the present study, the researches used melanoma cells and a RUNX2 knockout (RUNX2-KO) in vitro model to assess the influence of RUNX2 on CXCR4 protein levels along with its effects on markers associated with cell invasion and autophagy.
The Colo 800, Colo 853, Colo 679, A375, and MELHO cell lines expressed RUNX2 (Fig. 2a), the MMP13 invasiveness marker (Fig. 2b), and CXCR4 (Fig. 2c), at the messenger RNA (mRNA) level. The expression of RUNX2, MMP13 and CXCR4 at the protein level was also verified in the same melanoma cell lines (Fig. 2d, e).
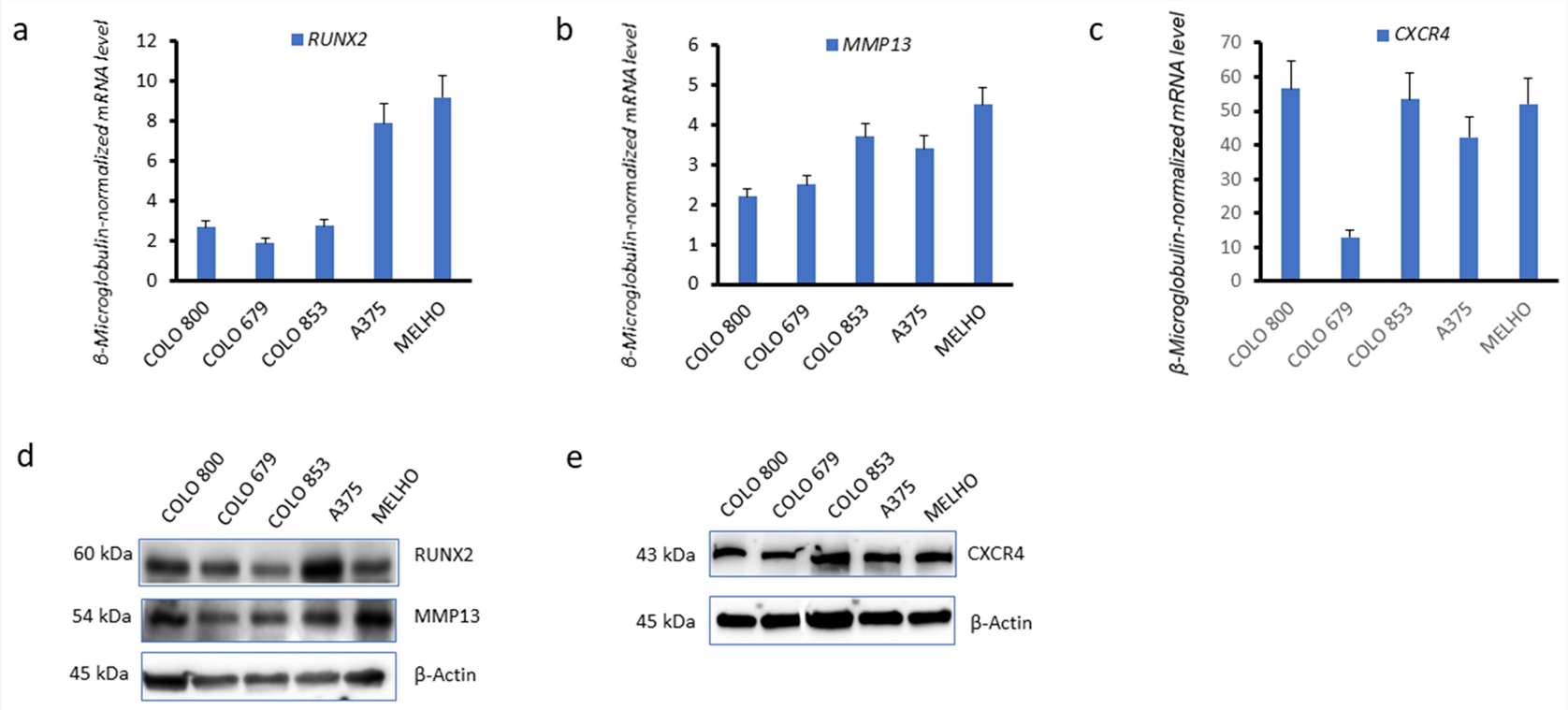 Fig. 2. RUNX2, CXCR4, and MMP13 are expressed in melanoma cell lines (Dalle Carbonare, Luca, et al. 2024).
Fig. 2. RUNX2, CXCR4, and MMP13 are expressed in melanoma cell lines (Dalle Carbonare, Luca, et al. 2024).
Western blotting analysis confirmed that the RUNX2 protein was not expressed in any of the three KO cell lines (1E7, 1B3, and 1F5) (Fig. 3a). Immunofluorescence further confirmed this observation in the three RUNX2 KO cell lines (Fig. 3b). Strikingly, real-time quantitative PCR (Fig. 3c), as well as Western blotting analysis (Fig. 3d), revealed that CXCR4 was downregulated in all of the three RUNX2 knockout cell lines. This observation indicated that RUNX2 is involved in CXCR4 expression.
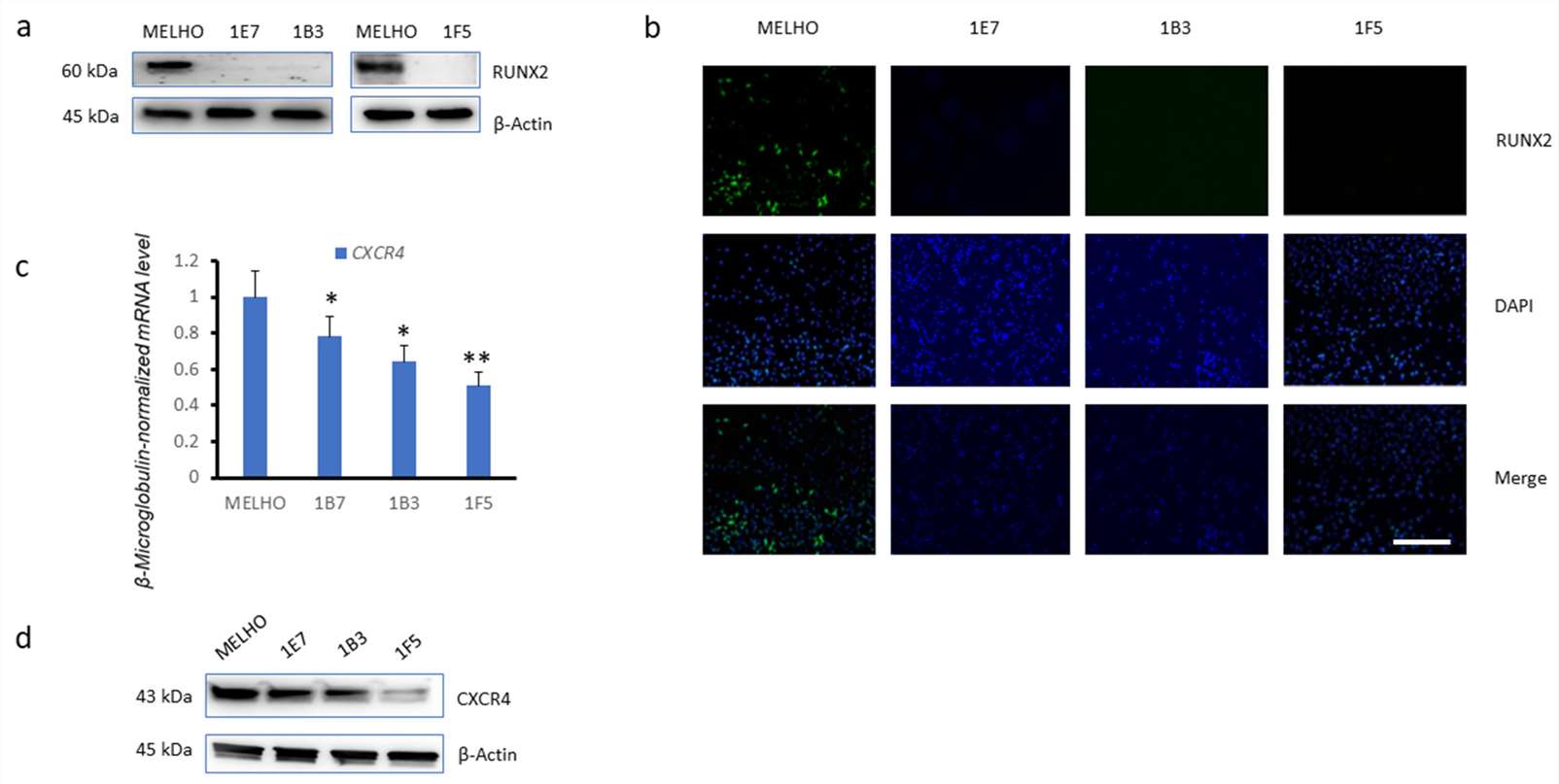 Fig. 3. Reduced expression of CXCR4 in RUNX2 knockout melanoma cell lines (Dalle Carbonare, Luca, et al. 2024).
Fig. 3. Reduced expression of CXCR4 in RUNX2 knockout melanoma cell lines (Dalle Carbonare, Luca, et al. 2024).
Ask a Question
Write your own review
- You May Also Need