Featured Products
Our Promise to You
Guaranteed product quality, expert customer support

Lung Tumor Cells
- Product List
- Background
- Applications
- Scientific Data
- FAQ
Lung cancer remains one of the most prevalent and deadly forms of cancer worldwide. Lung tumor cells originate from the epithelial cells lining the airways. These cells undergo genetic mutations that lead to uncontrolled growth, resulting in tumors. There are two main types of lung cancer: small-cell lung cancer (SCLC) and non-SCLC (NSCLC), each with distinct biological features and clinical behaviors. NSCLC is further categorized into adenocarcinoma, squamous cell carcinoma, and large cell carcinoma.
Lung tumor cells are characterized by a range of morphological features that distinguish them from their healthy counterparts. They often exhibit larger cell sizes, irregular shapes, and prominent, irregularly shaped nuclei, which are hallmarks of their rapid and uncontrolled proliferation. Additionally, lung tumor cells can display a high degree of cellular heterogeneity, with varying levels of differentiation and distinct subpopulations within the same tumor.
Genetic and Molecular Alterations in Lung Tumor Cells
- Gene mutations. Mutations in genes such as TP53, KRAS, and EGFR are commonly found in lung tumor cells. These mutations lead to the activation of oncogenes and the inactivation of tumor suppressor genes, promoting tumor growth and survival.
- Epigenetic changes. Epigenetic modifications, including DNA methylation and histone modification, play a significant role in lung cancer development. These changes can silence tumor suppressor genes and promote tumor growth.
- Chromosomal aberrations. Lung tumor cells often exhibit chromosomal rearrangements and numerical abnormalities. For instance, the translocation of the ALK gene is associated with a subset of lung adenocarcinomas, representing a target for specific inhibitors.
- Protein expression changes. Altered protein expression in lung tumor cells can affect various cellular processes, including cell cycle regulation, apoptosis, and angiogenesis.
In Vitro Modeling of Lung Cancer
Lung tumor cell lines are widely used to create in vitro models that mimic the tumor microenvironment. These models allow researchers to study the interaction between cancer cells and surrounding stromal cells, which is vital for understanding tumor growth, invasion, and metastasis. The ability to replicate the complexity of lung cancer in a dish makes it possible to test the effectiveness of potential therapies under controlled conditions.
High-Throughput Screening
The use of lung tumor cells in high-throughput screening (HTS) assays has accelerated the identification of novel therapeutic compounds. By exposing large libraries of compounds to lung tumor cells, researchers can quickly determine which substances have the potential to inhibit tumor cell growth or induce cell death. This approach is particularly valuable for discovering new drug candidates that can be further developed for clinical use.
Drug Resistance and Sensitivity
Lung tumor cells are used to investigate the mechanisms of drug resistance and to identify biomarkers that can predict patient response to specific treatments. This research can lead to more effective combination therapies and personalized treatment plans.
Gene Therapy and RNA Interference
Lung tumor cells are essential for developing gene therapy approaches, where genes are introduced, edited, or silenced to treat or prevent disease. RNA interference (RNAi) techniques, which use small RNA molecules to silence genes, are tested on lung tumor cells to identify potential therapeutic targets and develop novel treatments.
Glut1KO Neutrophils Negatively Impact Tumor Growth
Because the pro- and anti-tumor properties of neutrophils require a better understanding, the consequences of Glut1KO tumor-associated neutrophils (TAN) on tumor progression (Fig. 1A) were explored. Tumors with Glut1KO TANs had reduced cell proliferation as measured by phospho-Histone H3 (pHH3) and a reduced phospho-ERK1/2 (pERK) staining, a marker of tumor progression (Fig. 1B-C). Moreover, tumor growth rates monitored by longitudinal micro-computed tomography (μCT) were significantly diminished in mice with Glut1KO TANs (Fig. 1D). Next, to determine if TANs affect lung tumor cells in vitro, their behavior was measured using a KP-derived cell line (SV2) cultured alone, with control TANs or with Glut1KO TANs. A stronger negative impact on tumor cell growth was observed in Glut1KO TANs, as revealed by a reduced spreading of co-cultured SV2 cells (Fig. 1E). This suggests that Glut1KO TANs have a reduced tumor-supportive capacity or are endowed with anti-tumor properties.
To test if the reduced tumor-supportive function of Glut1KO TANs can be extended to other models, B16-F1 melanoma cells were injected via the tail vein of control or Glut1KO syngeneic recipient mice and the number of lung lesions was counted three weeks later. In this experimental metastasis model, there were fewer lesions in Glut1KO conditions (Fig. 1F). Furthermore, tumor cell proliferation was reduced in these tumors compared to control tumors, as indicated by a reduced proportion of pHH3-positive cells (Fig. 1G). Thus, TAN-mediated tumor support relies on Glut1 in different cancer types.
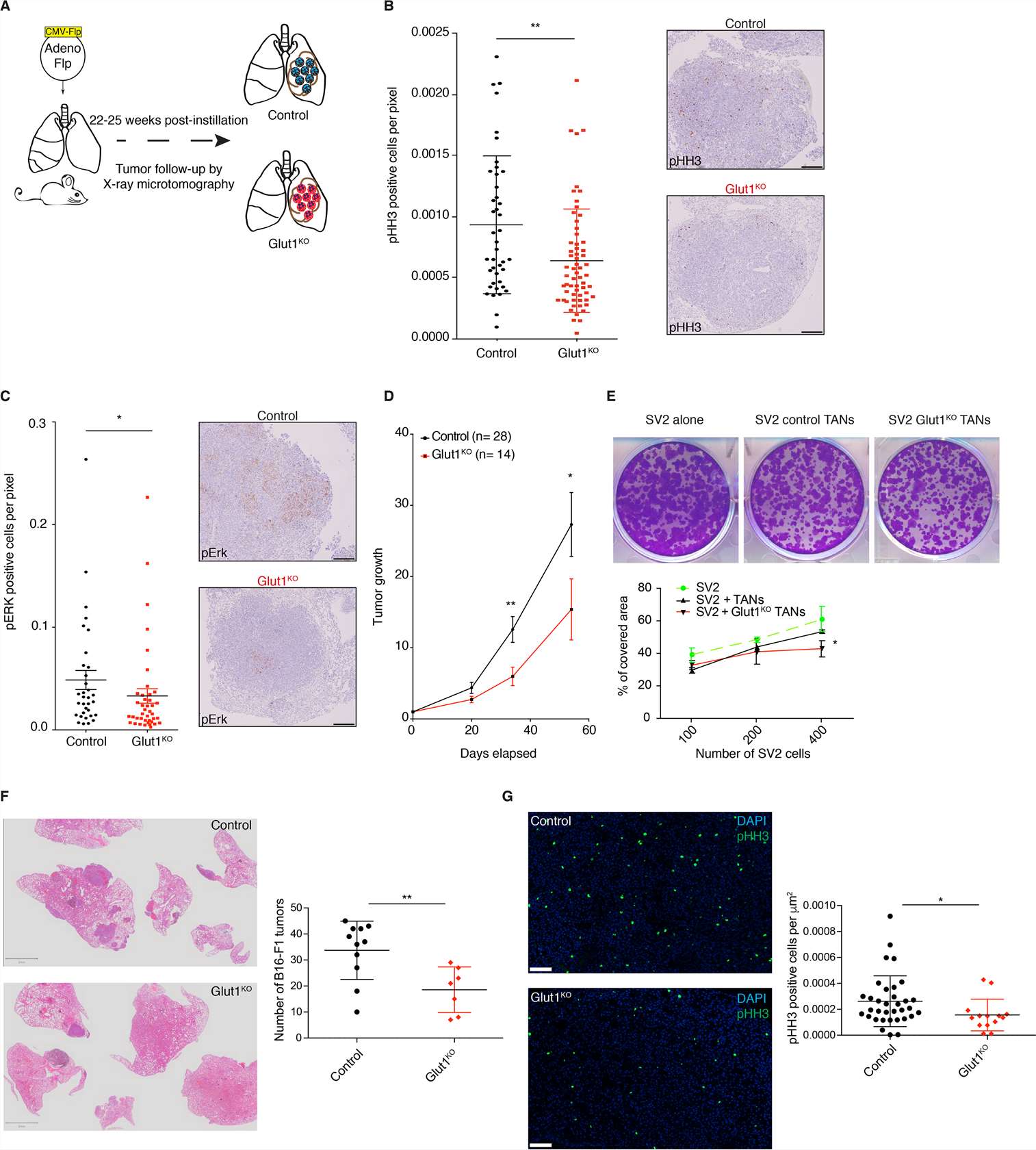 Fig. 1 Glut1 deletion in neutrophils reduces tumor growth. (Ancey PB, et al, 2021)
Fig. 1 Glut1 deletion in neutrophils reduces tumor growth. (Ancey PB, et al, 2021)
Secretion of EpCAM+ Lung Tumor Cell-Derived Exosomes is p53-dependent
To test the hypothesis that lung tumor cell-derived exosomes re-direct macrophage polarization, exosomes were isolated from conditioned media obtained from adenocarcinoma human alveolar basal epithelial cells (A549) and p53 null human lung cancer cells (H358) including the non-tumor epithelial cell line (HEK293) as a negative control. NanoSight analysis of isolated exosomes confirmed the particle size under a range of 50–200 nm (Fig. 2A) consistent with previous observations. The H358 exosomes were significantly smaller than those isolated from A549 (Fig. 2B). NanoSight analysis also showed a significantly lower concentration of H358 exosomes compared to A549 and HEK293 exosomes isolated from an equal volume of conditioned media (Fig. 2C), suggesting that as reported previously, p53 may regulate exosomes secretion from lung tumor cells. Further ImageStream analysis revealed the expression of epithelial cell adhesion molecule (EpCAM) and exosome-specific surface markers, including CD9, CD63, tumor susceptibility gene 101 (TSG-101), and CD81 (Fig. 2D, E) confirming the tumor cell-derived vesicles as exosomes.
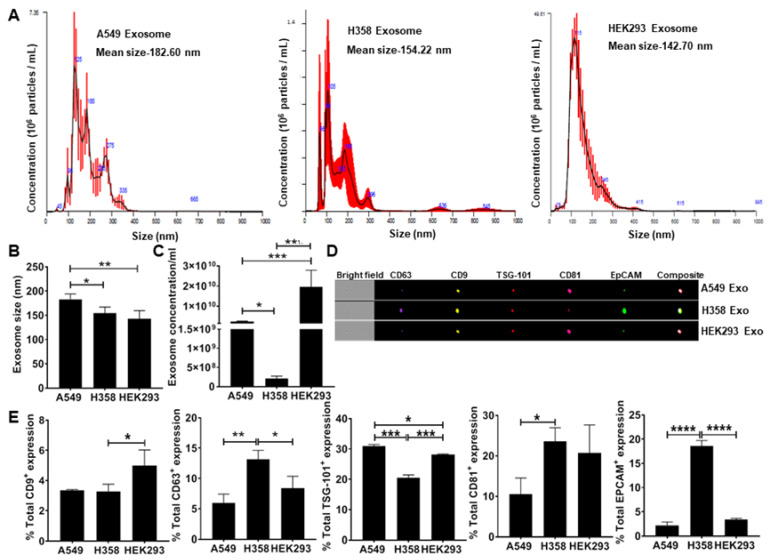 Fig. 2 Quantitation and characterization of tumor-cell derived exosomes. NanoSight and ImageStream analyses of A549-derived exosomes determined size, concentration, and characterization. Human lung cancer cells A549, H358, and non-tumor cells HEK293 were cultured in exosome-depleted media for 24 h. (Pritchard A, et al., 2020)
Fig. 2 Quantitation and characterization of tumor-cell derived exosomes. NanoSight and ImageStream analyses of A549-derived exosomes determined size, concentration, and characterization. Human lung cancer cells A549, H358, and non-tumor cells HEK293 were cultured in exosome-depleted media for 24 h. (Pritchard A, et al., 2020)
Lung tumor cells are abnormal cells that have undergone uncontrolled growth and division within the lungs. These cells can form solid masses or tumors that can be cancerous (malignant) or non-cancerous (benign).
The two main types of lung tumor cells are NSCLC and SCLC cells. NSCLC is the most common type, including adenocarcinoma, squamous cell carcinoma, and large cell carcinoma. SCLC is a less common but more aggressive type of lung cancer.
They divide and grow uncontrollably, forming a mass or tumor, which may have genetic mutations that allow them to avoid normal cellular growth control mechanisms.
Description: Human lung squamous cell carcinoma with PTHrP expression.
Description: established from the lymph node metastasis of a 59-year-old white male with lung adenocarcinoma
Description: established from the lung of a 72-year-old Japanese woman with lung adenocarcinoma
Description: established from the lung tissue of a 55-year-old female patient with lung adenocarcinoma
Description: established from the lung of a 55-year-old female patient with lung adenocarcinoma
Description: Isolated from the liver of a 44 year-old male patient at autopsy who had been treated...
Description: The HBE135-E6E7 cell line was derived from normal bronchial epithelium taken from...
Description: Established from a 25-year-old Caucasian male. The patient had received prior therapy...
Description: Species: human - female, fetus, 16 weeks old, Caucasian
Isoenzyme: G6PD,B
Histopathology: normal
Description: Species: human - embryo
Virus Susceptibility: poliovirus 1,2,3; adenovirus 3; vesicular...
Description: Species: human - male, 5 years old, Caucasian
Isoenzyme: G6PD, B
Histopathology: normal
Description: Species: human - female, 26 years old, Caucasian
Histopathology: pulmonary fibrosis
Description: Species: human - male, 18 years old, Caucasian
Isoenzyme: G6PD, B
Histopathology: normal
Description: COR-L23/5010 has been derived from the parent line, COR-L23 by continuous exposure...

