Laboratory Techniques to Detect Chromosomal Abnormalities in Multiple Myeloma
Blood Reviews. 2024 Mar; 64: 101168.
Authors: Clarke SE, Fuller KA, Erber WN
INTRODUCTION
Multiple myeloma is a plasma cell neoplasm driven by primary (e.g. hyper diploidy; IGH translocations) and secondary (e.g. 1q21 gains/amplifications; del(17p); MYC translocations) chromosomal events. These are important to detect as they influence prognosis, therapeutic response, and disease survival. Cytogenetic testing is commonly performed by interphase fluorescence in situ hybridization (FISH) on aspirated bone marrow samples. Several variations to the FISH methodology are available, including prior plasma cell enrichment and incorporation of immunophenotypic plasma cell identification. Other molecular methods are increasingly being utilized to provide a genome-wide view at high resolution (e.g., single nucleotide polymorphism (SNP) microarray analysis), which can detect abnormalities in most cases.
Karyotyping
Karyotyping (also known as "conventional") analysis is typically performed on a minimum of 20 G-banded metaphase spreads obtained after short-term culture of bone marrow aspirate specimens. It provides a full genome view of the cell and therefore can be used to identify structural and numerical abnormalities, ploidy status (such as hypodiploid karyotypes), as well as the presence of clonal heterogeneity.
Conventional karyotyping analysis is dependent on the presence of dividing cells. In myeloma, only 1-3% of neoplastic plasma cells are in the cell cycle, which limits the utility of detecting chromosomal defects. However, karyotypic abnormalities are identified in approximately 30% of cases; this may be enhanced with the addition of cytokines such as interleukin-4 to the culture conditions. Aberrant karyotypes are typically detected in more proliferative, advanced-stage diseases; consequently, their presence is considered more likely to be a surrogate marker for progressive disease and poor outcomes. Although deletion 13q/monosomy 13 was previously linked to poor prognosis when identified by karyotyping analysis, this is now believed to be more likely due to its link with advanced proliferative disease state as well as association with other high-risk markers such as hypodiploidy and t(4;14) IGH::FGFR3/MMSET. Some chromosomal defects are cryptic and cannot be delineated by G-banding (e.g. t(4;14) and t(14;16) translocations) since they involve small terminal portions of chromosomal material.
Fluorescence in Situ Hybridization
FISH is the "gold-standard" methodology for the assessment of chromosomal abnormalities in multiple myeloma and can be performed on cell suspensions from cultured bone marrow specimens or cells in interphase. FISH uses fluorescently labeled DNA probes to target specific chromosomal locations within the cell nucleus and is performed on cell preparations on glass slides. Metaphases are rarely used due to the limitations described above. Interphase FISH does not rely on the presence of dividing cells and can detect cryptic rearrangements, thus overcoming some of the deficiencies associated with karyotyping. Probes are available to detect prognostically relevant cytogenetic abnormalities and include dual fusion and break-apart probes for translocations and numerical and deletion probes to detect gains and losses of genetic material. A detailed description will be given of interphase FISH, its applications, and challenges.
- Sample preparation. The percentage of plasma cells in the marrow sample to be analyzed can be highly variable. This may be biological (i.e. disease burden in the marrow) or technical (i.e. quality or volume of the aspirate sample; haemodilution; first or subsequent "draw"). To address this, and to ensure there are sufficient plasma cells for cytogenetic analysis, many laboratories use plasma cell isolation or enrichment techniques before FISH processing; this is particularly used when there are<20% plasma cells in the sample. Plasma cell enrichment maybe by the removal of other hemopoietic cells (e.g. density gradient centrifugation) or plasma cell isolation (positive selection) techniques. The latter involves antigenic targeting of plasma cells using monoclonal antibodies (e.g. CD138), and immunomagnetic cell isolation or fluorescence-activated cell sorting (FACS). Immunomagnetic cell sorting (IMCS), which employs anti-CD138 antibodies to select plasma cells, is the enrichment technique most commonly utilized. FACS is more expensive but does enable the inclusion of additional markers to optimize plasma cell purity as well as address low plasma cell burden samples.
- Interpretation. To assess for deletions or gains of chromosomal material, between 100 and 200 interphase nuclei are scored for the number of signals present, while the presence of translocations is determined by enumerating colocalized signals (Fig. 1). Each probe type is associated with several different signal configurations: (i) "normal", (ii) "typical abnormal," representing the most frequently observed aberrations, and (iii) "variant abnormal" representing less frequently observed aberrations. In the setting of myeloma, variant signal patterns associated with clonal heterogeneity are often present, resulting in several signal patterns that need to be defined (Fig. 1).
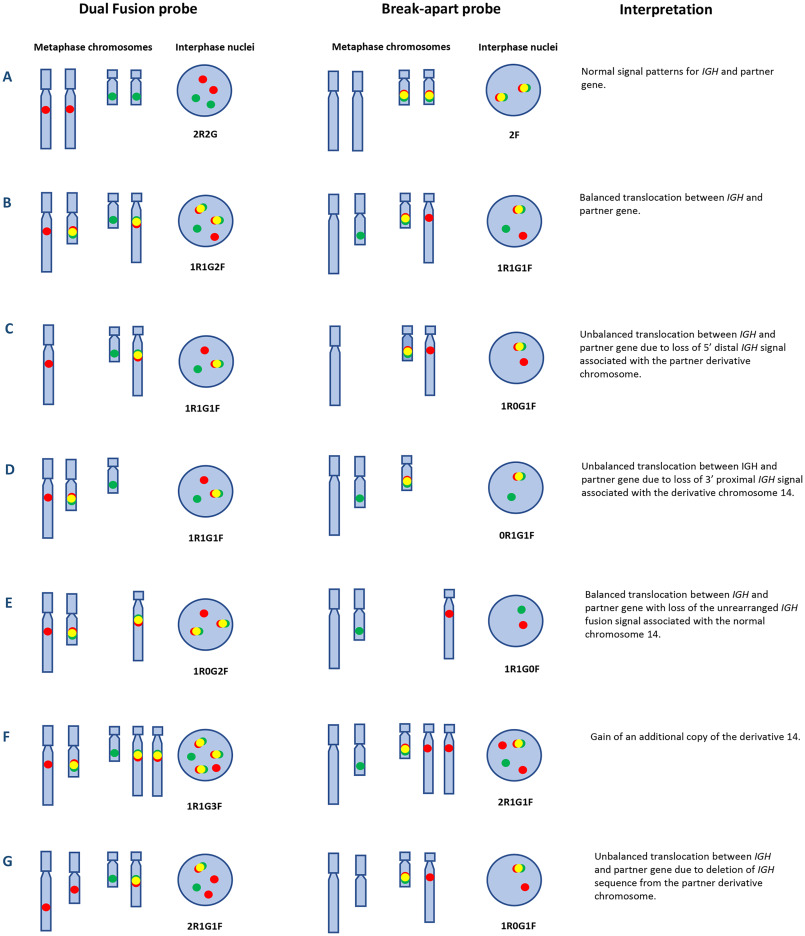 Fig. 1 Diagrammatic representation of normal and various abnormal IGH signal patterns in myeloma using IGH dual-fusion and break-apart probes.
Fig. 1 Diagrammatic representation of normal and various abnormal IGH signal patterns in myeloma using IGH dual-fusion and break-apart probes.
Other Fish-Based Methods
Other FISH-based methods involve positive plasma cell identification, rather than cell enrichment, leading to more targeted FISH analysis. These methods utilize morphological (i.e. "TargetFISH") or antigenic ("immuno") identification of plasma cells.
"Target FISH"
The "Target FISH" method uses automated imaging methodology to morphologically identify 100-200 plasma cells after density gradient enrichment and May-Grünwald Giemsa staining for comparison with FISH analysis. Slides are scanned and 100–200 plasma cells are identified. They are then de-stained with Carnoy's fixative, undergo FISH, and are rescanned to compare with the original morphological images. As this method is automated, it has the advantage of reduced user input; however, it is limited by the need for specific instrumentation and software.
ImmunoFISH
ImmunoFISH uses monoclonal antibodies to identify plasma cells based on antigen expression. This may be performed on cell spreads (e.g. cytocentrifuge preparations; bone marrow smears) or tissue sections (see below). Antibodies that are used include CD138 or immunoglobulin light chains of the paraprotein (also termed "cIg FISH") (Fig. 2). cIg FISH is more commonly used in clinical practice than CD138 cell identification as it can assist in distinguishing between tumor and normal plasma cells. The increased specificity enables the detection of significantly higher percentages of plasma cells harboring chromosomal abnormalities, and, when present, primary abnormalities are detected in almost all tumor cells. A dual enrichment and plasma cell identification approach can be used with initial density-gradient centrifugation to increase the plasma cell yield, cytocentrifugation, and antigenic identification; this is particularly helpful when there is a low disease burden in the sample. However, limitations of cIg FISH include increased training requirements for cytomorphologic analysis, false-negative results in non-secretory, non-producing myeloma cases, and difficulties in the detection of cases with atypical morphology (e.g. lymphoplasmacytic morphology associated with t(11;14)).
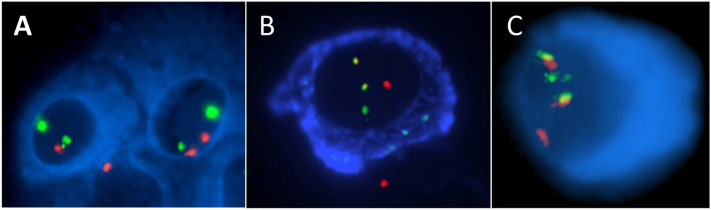 Fig. 2 Plasma cells analyzed by "immunoFISH" techniques.
Fig. 2 Plasma cells analyzed by "immunoFISH" techniques.
FISH on tissue sections
FISH can also be performed on formalin-fixed paraffin-embedded tissue sections of bone marrow or other involved extra-osseous tissues. This may be useful in cases where there is no suitable bone marrow aspirate sample, as it can be performed on bone marrow trephine biopsy. Although technically achievable, it is subject to difficulties associated with sample preparation and the ability to accurately identify the cells of interest. Tissue sections must be de-waxed and pre-treated with proteases to enable adequate access of the FISH probes to the genetic material. Bone marrow trephine sections must undergo de-calcification pre-treatment. This can cause various technical issues including reduced probe signal strength and high levels of autofluorescence. FISH signal enumeration can also be challenging due to tissue sectioning through nuclei, difficulties in distinguishing signals contained within tightly packed and overlapping nuclei, and evaluation of signals within different focal planes.
Flow Cytometry for Chromosome Detection
Flow cytometry can be used as an indirect measure of chromosome copy number or ploidy. Cells are incubated with DNA-binding fluorescent dyes (e.g. propidium iodide or DAPI) and the staining of cells in the G0G1 peak of plasma cells compared with normal cells. Achieving an accurate result commonly necessitates prior plasma cell enrichment. This is a surrogate measure only and does not give details of specific chromosomal alterations.
Imaging flow cytometry is a new sensitive and specific methodology that has been used to detect FISH abnormalities in myeloma. This uses flow cytometry principles, with the acquisition of thousands of cells, coupled with fluorescence microscopy. The method is highly specific as plasma cells are positively identified at the single cell level based on antigen expression (e.g. CD138 expression), analogous to immunoFISH and FICTION.
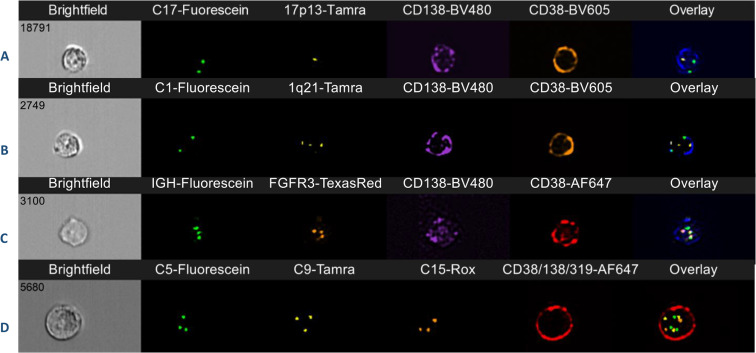 Fig. 3 Imaging flow cytometry of plasma cells in four cases of multiple myeloma.
Fig. 3 Imaging flow cytometry of plasma cells in four cases of multiple myeloma.
Creative Bioarray Relevant Recommendations
| Product/Service Types | Description |
| Human Tumor Cells | Creative Bioarray provides human tumor cells sourced from a variety of tissue types. Our human tumor cells have the original pathological diagnosis and are analyzed for key mutations, presenting the real characteristics of their in vivo state and maintaining heterogeneity across multiple passages, enabling you to expedite your drug discovery and process development. |
| Fluorescent In Situ Hybridization (FISH) Service | Creative Bioarray offers a range of different FISH services, including metaphase and interphase FISH (chromosomal assignment and clone ordering), fibre-FISH (chromosome painting), RNA-FISH (cell-based gene expression assay), M-FISH (multicolor karyotyping), 3D-FISH (on three-dimensionally preserved nuclei), flow-FISH (quantify the length of telomeres), immunoFISH (combined FISH and IHC). |
| Probe | Creative Bioarray provides the most comprehensive list of FISH probes for rapid identification of a wide range of chromosomal aberrations across the genome. |
RELATED PRODUCTS & SERVICES
Reference
- Clarke SE, et al. (2024). "Chromosomal defects in multiple myeloma." Blood Rev. 64: 101168.