Exploring the World of DART-FISH Technology
Nature Communications. 2024 Mar 20; 15 (1): 2511.
Authors: Kalhor K, Chen CJ, Lee HS, Cai M, Nafisi M, Que R, Palmer CR, Yuan Y, Zhang Y, Li X, Song J, Knoten A, Lake BB, Gaut JP, Keene CD, Lein E, Kharchenko PV, Chun J, Jain S, Fan JB, Zhang K.
INTRODUCTION
In situ transcriptomic techniques promise a holistic view of tissue organization and cell-cell interactions. There has been a surge of multiplexed RNA in situ mapping techniques but their application to human tissues has been limited due to their large size, general lower tissue quality, and high autofluorescence. Decoding Amplified taRgeted Transcripts with Fluorescence in situ Hybridization (DART-FISH) was reported, a padlock probe-based technology capable of profiling hundreds to thousands of genes in centimeter-sized human tissue sections.
DART-FISH Framework
DART-FISH involves in situ feature generation by padlock probe capture of targeted transcripts and rolling circle amplification (RCA), followed by a highly robust decoding process of sequential isothermal hybridization. Specifically, RNA molecules in fresh-frozen tissue sections are fixed with paraformaldehyde (PFA), permeabilized, and then reverse-transcribed with a mixture of random and poly-deoxythymidine (dT) primers. To assess the RNA content in human tissues as well as the retention of the cDNA molecules in situ, we added a 5' handle to the reverse-transcription primers to enable the collective visualization of all cDNA molecules with fluorescent oligos. We call this labeling method RiboSoma because the resulting signal labels the cell bodies. During protocol optimization, we noticed that crosslinking the cDNA molecules immediately after reverse-transcription to a polyacrylamide (PA) gel enhances the RiboSoma signal suggesting better retention of cDNA in situ throughout the DART-FISH protocol. This cDNA embedding strategy also led to a 1.5-fold median increase of the feature count per gene, compared to when the polyacrylamide gel is cast after RCA. Thus, RiboSoma serves as a marker for the cDNA content of the tissue and provides quality control for in situ reactions.
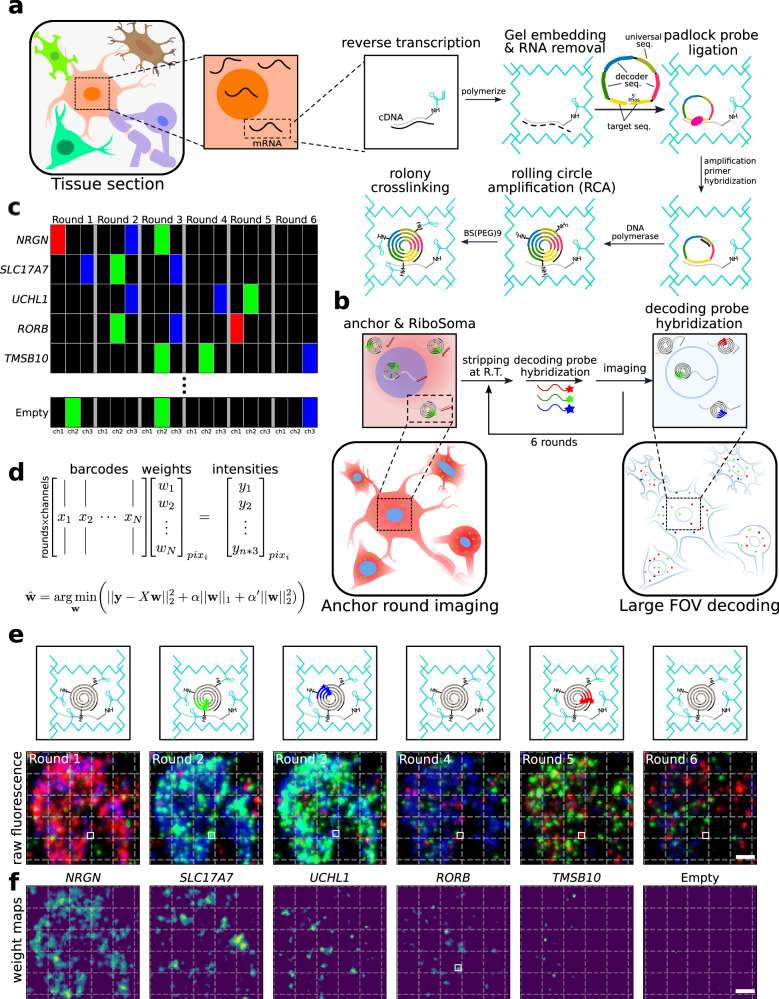 Fig. 1 DART-FISH workflow.
Fig. 1 DART-FISH workflow.
Following gel embedding and RNA digestion, cDNA molecules are hybridized with a library of padlock probes and circularized at a high temperature to ensure specificity. On their backbone, padlock probes carry a universal sequence used for amplification and gene-specific barcodes. The circularized padlock probes are then rolling-circle-amplified, generating RCA colonies in situ (rolonies) with hundreds of copies of barcode sequences concatenated in the form of a DNA nanoball. The rolonies are then covalently attached to the polyacrylamide gel to secure their positions during decoding. The result of the experiment is then assessed in the "anchor round" imaging, where fluorescent probes are hybridized to the universal sequences and the 5' handles on cDNA molecules to visualize the spatial distribution of all rolonies and cells.
Resolving Human Primary Motor Cortex Organizations With DART-FISH
To assess whether DART-FISH can resolve the organization of various cell types of human M1C, we set out to perform cell annotation by performing clustering on DART-FISH cells and matching them to the highest correlated subclass from a recent single-nucleus RNA sequencing (snRNA-seq) reference of M1C.
We resolved 20 subclasses from the major excitatory, inhibitory, and non-neuronal cell classes, which constituted 24.3%, 10.6%, and 65.1%, respectively, in the M1C. For excitatory neuronal subclasses, we successfully detected their laminar distribution, with L2/3 IT neurons localized at the superficial layer of the cortex and L6b/CT neurons deep in the cortex and close to the white matter, in line with the evolutionarily conserved organization of excitatory neurons in the mammalian M1C. Of note, L6 IT Car3 cells seem to be positioned more superficially than the L6 IT population, consistent with recent observations in the human visual cortex and middle temporal gyrus. In contrast, inhibitory neuronal subtypes generally showed wider spatial gradients along the cortical axis; for instance, the Vip population was enriched in layers 2-4 as suggested by previous studies in the mouse.
Moreover, we observed some cells belonging to the excitatory neurons and inhibitory neurons localized in the white matter region, which likely are the adult remnants of early generated subplate neurons discovered in previous studies. For non-neuronal cells, we observed oligodendrocytes appearing at layer 4 and peaking in the white matter despite the uniform distribution of the oligodendrocyte progenitors across the tissue section.
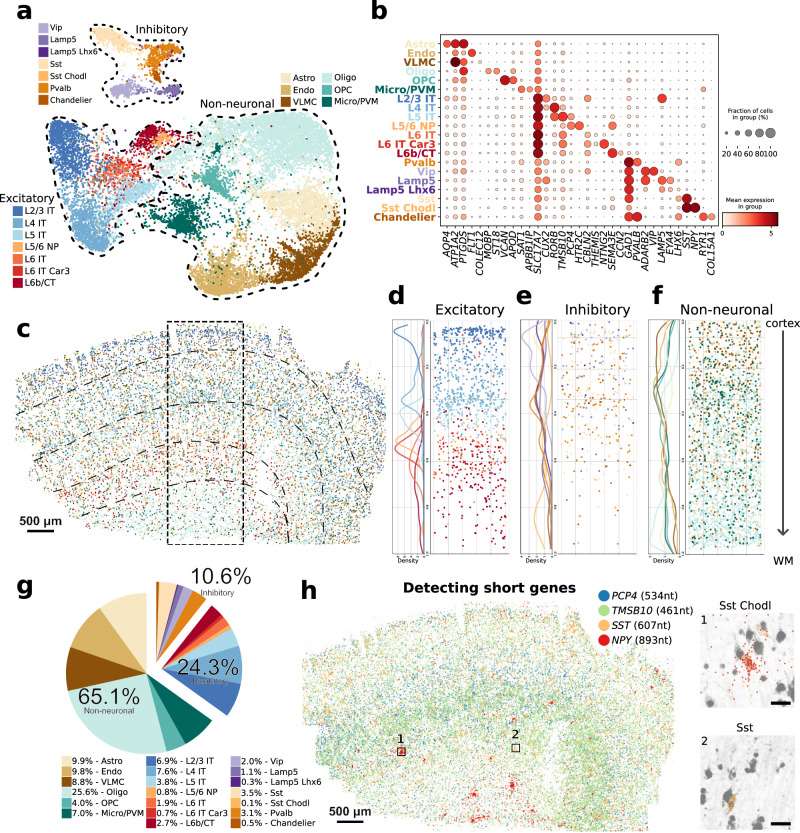 Fig. 2 DART-FISH mapping of cell types in the human M1C.
Fig. 2 DART-FISH mapping of cell types in the human M1C.
We further assessed whether we could detect short genes (<1.5kb) with DART-FISH. We compiled a list of 33 differentially expressed genes shorter than 1.5kb comprising well-studied genes as well as less well-known computationally derived marker genes in the brain. For example, by targeting SST (607 nt) and NPY (893 nt), we could uncover a rare subclass of inhibitory neurons, Sst Chodl (0.1% abundance), specified by the expression of these short neuropeptides.
Creative Bioarray Relevant Recommendations
| Product/Service Types | Description |
| In Situ Hybridization (ISH) & RNAscope Service | Creative Bioarray provides ISH services using fluorescence or bright fields. |
| Fluorescent In Situ Hybridization (FISH) Service | Creative Bioarray offers a full line of FISH services, from standardized testing of validated assays to custom development of new assays. |
| ISH/FISH Probes | Creative Bioarray provides the most comprehensive list of FISH probes for rapid identification of a wide range of chromosomal aberrations across the genome. |
| FISH Probe Design, Synthesis and Testing Service | With access to the entire RP11 BAC library, Creative Bioarray is capable of developing custom FISH probes. Apart from that, we also offer mRNA ISH/FISH probes, miRNA ISH/FISH probes, and lncRNA ISH/FISH probes. |
RELATED PRODUCTS & SERVICES
Reference
- Kalhor K, et al. (2024). "Mapping human tissues with highly multiplexed RNA in situ hybridization." Nat Commun. 15 (1): 2511.