Enrichment, Isolation and Characterization of Circulating Tumor Cells (CTCs)
Introduction
Circulating tumor cells (CTCs) are considered to be an important factor that takes part in the dissemination of metastases in patients with cancer. Current technologies allow for the isolation of CTCs from the peripheral blood of patients, which can be subsequently analyzed at the genetic and molecular level for the detection of tumor-associated genes and/or antigens.
CTCs are very rare and have a short half-life in the bloodstream. Usually, between 1 to 10 CTCs can be detected per milliliter of peripheral blood of patients with metastatic stage cancer. The ratio of CTCs and peripheral blood mononuclear cells may range between 1 to 105 and 1 to 106. For this reason, peripheral blood-derived CTCs fractions require an enrichment before they can be analyzed. Methods for enrichment and analysis are based on physical and/or biological properties of CTCs, such as size, deformability, density, polarity and electrical charge, epithelial cell adhesion molecule (EpCAM), cytokeratins (CKs) and tumor-associated markers expression.
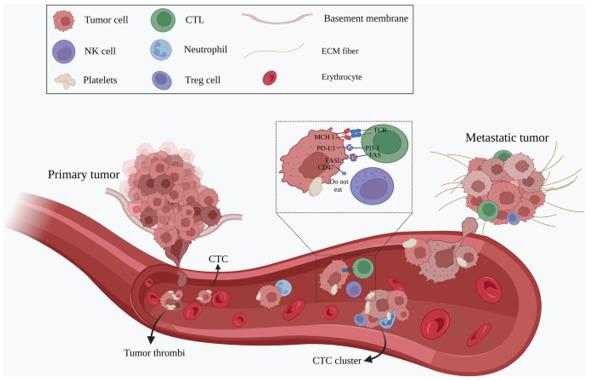 CTCs leave the primary tumor as single cells or in clusters, intravasate into the bloodstream and travel through the circulation to the distant site of the body to establish metastasis (Ju, S., Chen, C., Zhang, J. et al., 2022).
CTCs leave the primary tumor as single cells or in clusters, intravasate into the bloodstream and travel through the circulation to the distant site of the body to establish metastasis (Ju, S., Chen, C., Zhang, J. et al., 2022).
CTCs Enrichment
Density-dependent cell separation
This method for CTCs enrichment from peripheral blood utilizes an inert polysucrose termed Ficoll, which was originally produced for the isolation of mononuclear blood cells from whole blood. The Ficoll protocol relies on differential migration properties that derive from diverse cell-type dependent buoyant densities. A flaw of the density-based cell separation method is the possible loss of some CTCs.
Separation based on cell size
This type of CTCs enrichment is based on size and deformability of epithelial cancer cells. A 10-cm silicon wafer device was produced with standard micromachining techniques. This device consists of four segments of microfluidic channels placed on a silicon wafer, which are attached by adjacent little tanks that contain a fluid inlet and outlet. Another microdevice contains multiple arrays of crescent-shaped isolation wells for the isolation of epithelial cancer cells from blood cells. Each trap has a gap of 5μm, which allows for the exit of blood components, including larger white blood cells that are more deformable than malignant cells. Microdevices are engineered to enhance hydrodynamic efficiency and to facilitate the optimal retrieval of CTCs. A microfilter made of a parylene membrane was produced for the isolation, electrolysis and genomic analysis of CTCs. Size-based enrichment systems are limited by the significant variations in cell size among cancer cell populations, because of tumor heterogeneity. This may result in the loss of some CTCs.
Negative enrichment
Negative enrichment technique enriches CTCs by depleting most of the leukocytes and erythrocytes. This technique utilizes antibodies directed against hematopoietic cells and crosslink them to multiple erythrocytes, which in turn leads to the formation of immunorosettes. Centrifugation over the Ficoll-Paque buoyant density medium allows for the precipitation of immunorosettes and unbound red blood cells, while CTCs fractions can be recovered from the medium. This protocol is limited by the possible loss of some CTCs.
Negative enrichment by immunomagnetic beads
Ferromagnetic beads coated with anti-CD45 antibodies can be used as a further purification step for CTCs derived from density gradient centrifugation procedures. CD45 is a protein tyrosine phosphatase expressed on the membrane of hematopoietic cells, with the exception of erythrocytes and plasma cells. CD45 is also missing in epithelial cells. Thus, anti-CD45 immunomagnetic beads only bind to blood cells, which can be subsequently removed with a magnetic field. The capture efficiency of this protocol ranges from 52% to 88%. Limitations of this technique are the possible loss of some CTCs and the presence of impurities, as not all CD45-positive cells are removed during the procedure.
Positive enrichment by immunomagnetic beads
This method is also known as magnetic activated cell sorting (MACS). Ferromagnetic beads coated with anti-EpCAM antibodies can be used to isolate CTCs, following a density gradient centrifugation step. EpCAM-positive CTCs can be automatically isolated with the MagSweeper, which uses a magnetic arm to harvest the ferromagnetic beads. MACS-based CTCs isolation relies on the single target molecule used in the protocol. In addition, some CTCs with low EpCAM expression and/or CTCs that have undergone advanced epithelial-mesenchymal transition (EMT) might be lost.
Magnetophoretic mobility-based separation
In this negative enrichment cell sorting system, cells are labeled with magnetic nanoparticles. The Quadrupole Magnetic Cell Sorter (QMS) was used to rapidly enrich and isolate CTCs from hematopoietic cells. QMS can sort 10 million cells per second, with a 99% depletion efficiency of CD34+ and CD45+ peripheral blood cells. This system allows only for the depletion of unwanted cells. After the enrichment step, other detection techniques must be applied for the isolation of pure CTCs populations.
Microfluidic devices
In microfluidic devices, the blood flows on so-called CTC-chips, which are made of equilateral triangle structures containing an assortment of anti-EpCAM antibody-coated microposts or chips with other configurations such as graphene oxide chips. However, the enhanced sensitivity for capturing cell lines with low EpCAM expression might not be sufficient to select biopsy-derived CTCs that have undergone advanced EMT.
CTCs Detection
Nucleic acid-based systems
Quantitative RT-PCR is the preferred nucleic acid-based method for analyzing CTCs-related markers. Furthermore, studies showed that RT-PCR is more sensitive than immunohistochemistry for detecting cell specific markers. RT-PCR may be susceptible to provide false positives due to sample contamination. There is also the possibility that the target gene is expressed in some normal cells. Another disadvantage of RT-PCR is that analyzed cells cannot be used for any other type of study.
Fluorescence in situ hybridization (FISH)
This technique utilizes fluorescent labeled DNA probes to detect specific DNA sequences within certain chromosomes. FISH analysis is very precise and can detect a variety of genetic abnormalities associated with human diseases. However, the experimental procedure requires highly trained personnel, is labor intensive and sometimes may fail to provide clear results. The analyzed cells cannot be used for subsequent studies, as they are no longer viable after FISH analysis.
Integrating antigen-independent subtraction enrichment (SE) and immunostaining-FISH (SE-iFISH)
This technique relies on an enrichment step for CTCs, in which cells retain viability and are suitable for primary tumor cell culture conditions. The enrichment step is followed by immunostaining for epithelial markers, combined with FISH analysis for chromosomal aneuploidy. SE-iFISH technology can be supported by the Metafer-iFISH automated CTC imaging system to provide a platform for the characterization of each CTC subtype in clinical disease progression.
The iFISH method allows for the in situ phenotyping and karyotyping of CTCs, along with the classification of various CTCs subtypes, following the detection of tumor biomarkers expression levels.
Fluorescence assisted cell sorting (FACS)
FACS relies on the use of antibodies and is often used for the separation of a specific cell type, which can be isolated with high purity from the general cell population. Forward scatter and side scatter allow for the characterization of physical properties such as cell size and internal cellular complexity, respectively. An electrostatic detection system leads to the accurate isolation of a defined cell fraction, based on the electric charge of that particular cell population. FACS is a versatile technique with a wide range of applications, as several parameters can be analyzed simultaneously. FACS has a limited throughput, because cells are sorted individually. FACS technology may suffer from another shortcoming. In some cases, flow sorting conditions may be detrimental to particular types of cells and, therefore, impede subsequent studies.
Methods for the Simultaneous Enrichment and Detection of CTCs
Fiber-optic array scanning technology (FAST)
FAST cytometry was developed to speed up the analyses of immunofluorescent-labeled cells in large volumes of peripheral blood. A laser printing technique is used to localize rare subpopulations of immunofluorescent-labeled cells on glass substrates. The laser printing optic has the ability to excite 300,000 cells per second and the emission spectrum is monitored on a very wide field of view. The scan rates of FAST can be 500-fold faster than automated digital microscopy. The enhanced screening rate combined with the high sensitivity provides the desired performance for early detection of peripheral blood of early-stage tumors. The possibility to detect and isolate CTCs in a single step minimizes the chance of losing cells.
CellSearch
In CellSearch, a ferrofluid linked to anti-EpCAM antibodies is utilized for the initial enrichment of EpCAM-positive cells, and subsequently, a magnetic field is applied to isolate the ferrofluid-EpCAM-positive cell complex. CTCs detection is carried out with a set of phycoerythrin (PE)-labeled anti-CK antibodies and allophycocyanin (APC)-labeled anti-CD45 antibodies, mixed with a nuclear dye (DAPI) and a permeabilization buffer for the entry of anti-cytokeratin antibodies into the cells. Finally, immunofluorescence is carried out for the enumeration of EpCAM-positive CK-positive CD45-negative CTCs.
The CellSearch system can only detect EpCAM-positive and CK-positive CTCs. This system is not suitable for CTCs that either do not express epithelial phenotypes, or have lost EpCAM and CK expression, due to advanced EMT.
ISET (isolation by size of epithelial tumor cells)
ISET enriches epithelial cancer cells with a blood filtration, which takes place through a membrane that has pores with a diameter of 8 μm. Larger epithelial cancer cells remain on the filter and, therefore, can be stained either for immunocytochemistry, or cytomorphological analysis. ISET is capable of detecting a single CTC from 1 ml of peripheral blood.
A comparative analysis shows some discrepancies in the enumeration of CTCs between ISET and CellSearch. These discrepancies may be attributable to the heterogeneity of tumor cell size and/or to differential expression of EpCAM in cancer cells.
AdnaTest
In AdnaTest, CTCs enrichment is conducted with magnetic separation of EpCAM-positive cells, whereas the detection is carried out by RT-PCR. Both AdnaTest and CellSearch exhibited concordant data in terms of CTCs enumeration and sensitivity. Both systems were able to detect 2 CTCs in 7.5 ml of patient-derived peripheral blood. However, the cells are no longer viable after AdnaTest, and therefore, they cannot be used for other studies. Additionally, RT-PCR technology may provide false positives due to nucleic acid-related contaminations.
EPISPOT
EPISPOT detects specific secreted proteins and stands for epithelial immunospot, which derives from enzyme-linked immuno-assays. An initial enrichment step is carried out by immunomagnetic depletion of CD45-positive cells, which is then followed by immunomagnetic positive enrichment by selecting CXCR4-positive cells. This system is restricted to the analysis of CXCR4-positive CTCs, so CXCR4-negative CTCs are lost.
Ariol system
The Ariol system is commercially available and consists of an automated cell image capture for the analysis of CTCs placed on glass slides. For the enrichment step, peripheral blood is initially treated with a lysis buffer to remove erythrocytes. Cell blocking, permeation and fixation were then carried out. Following the instructions of the CTC enrichment and detection kit to perform CTCs enrichment with anti-CK antibodies either alone, or in combination with anti-EpCAM antibodies. The Ariol system allows detection of CK-positive EpCAM-positive and CK-positive EpCAM-negative CTCs. However, cells are not viable after the Ariol system assay.
Creative Bioarray Relevant Recommendations
| Products & Services | Description |
| SuperBeads® Human CTC Isolation Kit (Direct) | Use SuperBeads® Human CTC Isolation Kit (Direct) to isolate CTCs directly from human whole blood by immunomagnetic negative selection. The kit depletes other kinds of cells. The negatively isolated Human CTCs are left in the sample and have not been touched with the SuperBeads. |
| Circulating Tumor Cell (CTC) FISH | Creative Bioarray now offers a full line of circulating tumor cell fluorescence in situ hybridization (FISH) services, from enrichment of CTCs, standardized testing of validated assays, and custom development of new assays. |

