Comparison of IHC, NGS, and FISH for Detection of MTAP Loss
Modern Pathology. 2024 Mar; 37 (3): 100420.
Authors: Febres-Aldana CA, Chang JC, Jungbluth AA, Adusumilli PS, Bodd FM, Frosina D, Geronimo JA, Hernandez E, Irawan H, Offin MD, Rekhtman N, Travis WD, Vanderbilt C, Zauderer MG, Zhang Y, Ladanyi M, Yang SR, Sauter JL.
INTRODUCTION
9p21 deletions involving MTAP/CDKN2A genes are detected in diffuse pleural mesotheliomas (DPM) but are absent in benign mesothelial proliferations. Loss of MTAP expression by immunohistochemistry (IHC) is well accepted as a surrogate for 9p21 deletion to support a diagnosis of DPM. Accurate interpretation can be critical in the diagnosis of DPM, but variations in antibody performance may impact interpretation. The objectives of this study were to compare the performance of MTAP monoclonal antibodies (mAbs) EPR6893 and 1813 and to compare MTAP expression by IHC with 9p21 copy number status in DPM.
METHODS
Specimens included in the study comprised consecutive clinical samples of DPM diagnosed at the study institution between January 2014 and January 2021, with tissue available for IHC and prior clinical next-generation sequencing (NGS) data available.
- All DPM specimens underwent targeted NGS by MSK-IMPACT to serve as a reference for 9p21 locus status before performing MTAP IHC on each tumor. Fraction and allele-specific copy number estimates from tumor sequencing (FACETS) perform a systematic enumeration of allelic-specific read counts from all heterozygous and homozygous SNP loci as well as pseudo-SNPs (nonpolymorphic loci) along target intervals. Integer copy number calls of major and minor alleles of 9p21 were reviewed to distinguish copy number states with the following criteria: diploid state (2,1), loss of heterozygosity (LOH) (1,0), copy number neutral LOH (≥ 2, 0), and homozygous deletion (0,0).
- MTAP IHC was performed on formalin-fixed paraffin-embedded (FFPE) tissue (from the same tissue block used for NGS in all cases) using MTAP mAb clone EPR6893 at 1:1000 dilution (0.9 μg/mL) and MTAP mAb clone 1813 at 1:2500 (0.2 μg/mL). Positive and negative staining with both antibodies was evaluated with appropriate control tissue.
- FFPE tissue sections of 4-μm thickness with tumor areas marked were used for FISH analysis following standard protocols. CDKN2A (9p21) FISH probe of 222 kb, labeled in orange, was used in combination with a centromere-specific probe for chromosome 9 (CEP9), labeled in green. Signal analysis was performed in combination with morphology correlation, with at least 50 interphase cells within the marked tumor area evaluated. The selection of tumor cells for analysis was primarily on those with at least 2 internal control signals of the CEP9 probe. Homozygous deletion of the CDKN2A gene was recorded if cells showed no signals for the CDKN2A gene and 2 or more signals for the internal control probe CEP9.
- Browse our recommendations
| Product/Service Types | Description |
| FFPE Cell Pellet | Creative Bioarray has also established standard operation procedures in custom cell pellets and cell pellet arrays. We offer clients the opportunity to customize their requests to meet their unique needs. |
| Immunohistochemistry (IHC), Immunofluorescence (IF) Service | Creative Bioarray offers a comprehensive IHC service from project design, and marker selection to image completion and data analysis. We are dedicated to satisfying every customer and assisting them to achieve their specific research goals. |
| Fluorescent In Situ Hybridization (FISH) Service | Creative Bioarray offers a range of different FISH services, including metaphase and interphase FISH (chromosomal assignment and clone ordering), fibre-FISH (chromosome painting), RNA-FISH (cell-based gene expression assay), M-FISH (multicolor karyotyping), 3D-FISH (on three-dimensionally preserved nuclei), flow-FISH (quantify the length of telomeres), immunoFISH (combined FISH and IHC). |
RESULTS
- Fifty-six DPMs were identified for IHC and NGS profiling. NGS profiling of 56 DPM revealed a median of 2 somatic mutations per sample. The median sample coverage was 633x (range: 207-956x) and the median tumor purity was 30% (range, 10%-85%) with 38 cases (68%) showing tumor purity of greater than 20%. Overall, copy number analysis using FACETS demonstrated homozygous deletion of 9p21 in 25 (45%) DPM.
- Of 56 DPM tested with mAb EPR6893, 19 cases (34%) showed equivocal interpretations, whereas 17 (30%) and 20 (36%) DPM exhibited retained and loss of cytoplasmic MTAP expression, respectively. Distribution of cases with equivocal, retained, and loss of MTAP expression by histologic subtype included 16, 16, and 15 epithelioid DPM; 2, 1, and 4 biphasic DPM; and 1, 0, and 1 sarcomatoid DPM, respectively. MTAP expression (whether equivocal, retained, or lost) in biphasic tumors was the same in both epithelioid and sarcomatoid components. Equivocal interpretations included weak "blush" cytoplasmic staining in 10 cases, poor internal control in 6 cases, and heterogeneous staining in 3 cases.
- There were no equivocal interpretations in all 56 cases of DPM tested with mAb 1813. Twenty-one cases (37%) showed retained cytoplasmic MTAP expression in tumor cells with mAb1813, whereas 35 (63%) exhibited MTAP loss in tumor cells. MTAP expression (whether retained or lost) in biphasic tumors was the same in both epithelioid and sarcomatoid components. One case with MTAP loss by IHC and homozygous deletion by FACETS demonstrated a subclonal immunohistochemical staining pattern with loss of MTAP expression restricted to a subset of tumor cells. Distribution of cases with retained and loss of MTAP expression by histologic subtype included 20 and 27 epithelioid DPM; 1 and 6 biphasic DPM; and 0 and 2 sarcomatoid DPM, respectively.
- There were no equivocal cases with mAb 1813. MTAP mAb 1813 showed stronger immunoreactivity overall, reducing nonspecific "blush" background staining often seen with mAb EPR6893, and improvement in the intensity of staining of positive internal control, facilitating better distinction in interpreting loss or retained expression (Fig. 1 and 2). Fifteen (79%) and 4 (21%) cases that were equivocal with mAb EPR6893 showed loss and retained MTAP expression with mAb 1813, respectively. The case with subclonal loss of MTAP expression with mAb 1813 was an epithelioid DPM, initially interpreted as equivocal with mAb EPR6893 (Fig. 2A, a).
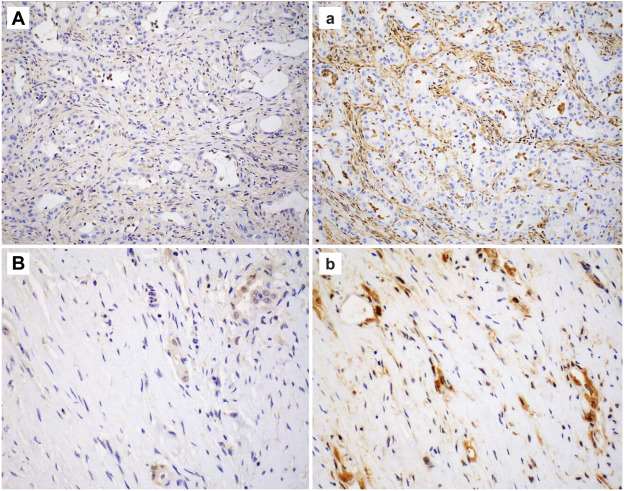 Fig. 1 MTAP immunoreactivity using mAb EPR6893 (uppercase A, B) compared with mAb 1813 (lowercase a, b).
Fig. 1 MTAP immunoreactivity using mAb EPR6893 (uppercase A, B) compared with mAb 1813 (lowercase a, b).
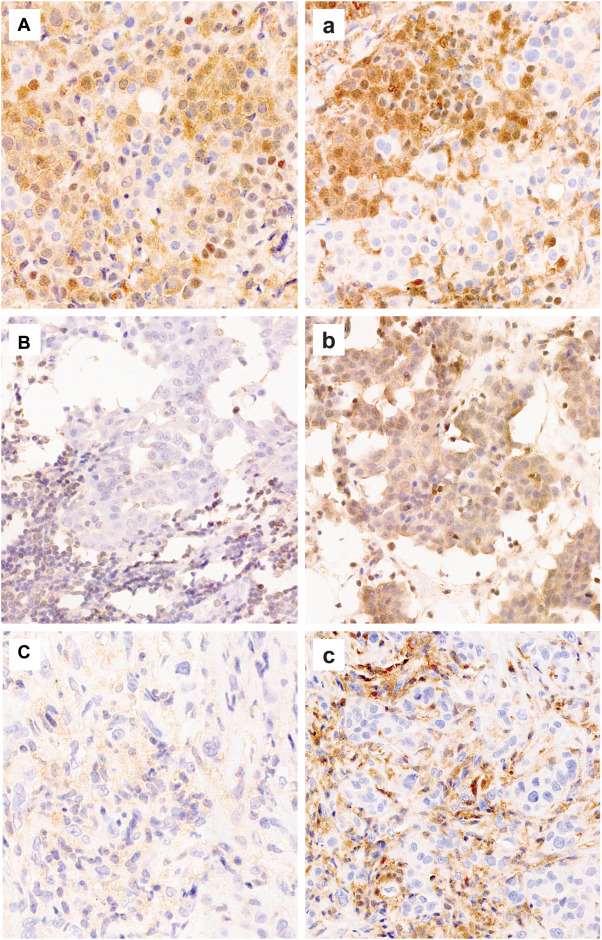 Fig. 2 MTAP immunoreactivity using mAb EPR6893 (uppercase A-C) compared with mAb 1813 (lowercase a-c).
Fig. 2 MTAP immunoreactivity using mAb EPR6893 (uppercase A-C) compared with mAb 1813 (lowercase a-c).
- Results of 9p21 copy number analyses using FACETS and FISH are shown in Figure 3. Copy number by FACETS was analyzed for all samples, including 38 cases (68%) with greater than 20% tumor purity and 18 cases (32%) with tumor purity of 20% or less, which is the recommended minimum tumor purity for optimal performance. 32 (91%) of 35 cases that showed MTAP loss by IHC with mAb 1813 showed concordant 9p21 homozygous deletion by either FACETS or CDKN2A FISH, whereas 3 (9%) were discordant with homozygous 9p21 deletion not detected by FACETS. In a single case with additional tissue available, FISH also confirmed the lack of CDKN2A homozygous deletion. Of 21 cases that showed retained MTAP expression by IHC with mAb 1813, 20 (95%) cases were concordant with no 9p21 deletion detected by FACETS, and 1 (5%) case was discordant with homozygous 9p21 deletion detected by FACETS. Unfortunately, additional tissue was not available in this case for orthogonal FISH testing to confirm the 9p21 deletion by FACETS (Fig. 3).
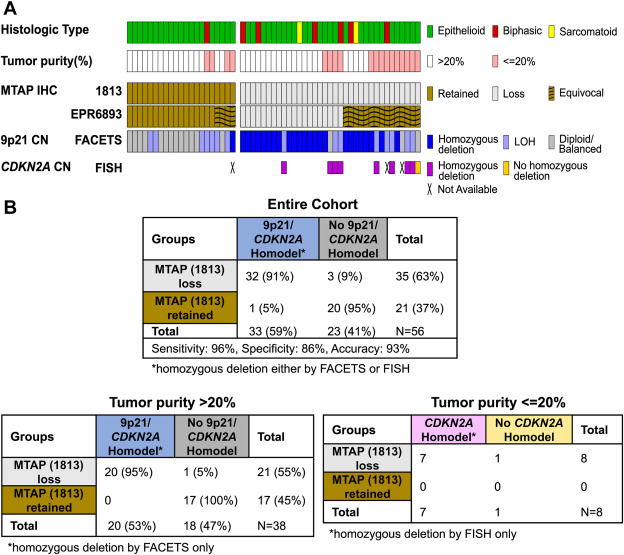 Fig. 3 Correlation of MTAP expression by IHC and 9p21 copy number by NGS and CDKN2A FISH.
Fig. 3 Correlation of MTAP expression by IHC and 9p21 copy number by NGS and CDKN2A FISH.
SUMMARY
Interpretation of MTAP IHC is improved with mAb 1813 because mAb EPR6893 was often limited by equivocal interpretations. MTAP IHC and molecular assays are complementary in detecting 9p21 homozygous deletion. MTAP IHC may be particularly useful for low tumor purity samples and in low-resource settings.
RELATED PRODUCTS & SERVICES
Reference
- Febres-Aldana CA, et al. (2024). "Comparison of Immunohistochemistry, Next-generation Sequencing and Fluorescence In Situ Hybridization for Detection of MTAP Loss in Pleural Mesothelioma." Mod Pathol. 37 (3): 100420.