B-Cell Tumors With t(14;19)(q32;q13)/IGH::BCL3 Identified by G-Banding and FISH
Journal of Clinical and Experimental Hematopathology. 2024; 64 (1): 21-31.
Authors: Ohno H, Maekawa F, Hayashida M, Nakagawa M, Fukutsuka K, Matsumura M, Takeoka K, Maruyama W, Ukyo N, Sumiyoshi S, Tanaka Y, Haga H.
INTRODUCTION
t(14;19)(q32;q13) was first described in 3 out of 30 patients with B-cell chronic lymphocytic leukemia (B-CLL), and the translocation was found to create a fusion gene between the IGH gene at the 14q32 chromosomal band and BCL3 gene at 19q13. In a large series of B-CLL patients tested by fluorescence in situ hybridization (FISH) using relevant probes, t(14;19)(q32;q13)/IGH::BCL3 was found in 16 (0.9%) out of a total of 1,683 patients.
METHODS
We searched the database of cytogenetic analysis in our institution between 2010 and 2023 and selected 5 patients who carried t(14;19)(q32;q13), including one patient described previously.
- Mononuclear cells were separated from peripheral blood (PB) or bone marrow (BM) aspirates. Lymph node (LN) biopsy specimens were aseptically minced to prepare a cell suspension. Cells were resuspended in phosphate-buffered saline and aliquots were subjected to flow cytometry (FCM).
- Pathological specimens were fixed in 10% neutral buffered formalin, embedded in paraffin, and then subjected to histopathological examination. The monoclonal antibodies used for immunohistochemistry (IHC) were: anti-CD3, CD5, CD10, CD20, CD 30, CD79a, BCL2, MYC, BCL3, and PD-1.
- Tumor cells prepared from PB/BM/LN were incubated in RPMI 1640 medium supplemented with 15% heat-inactivated fetal bovine serum at 37°C under a CO2 concentration of 5%. Cells were then cultured in the presence of 0.1 μg/mL colcemid for 2 hr. After harvesting, the cells were treated with the hypotonic solution and fixed in methanol: acetic acid (3:1). Chromosomes were banded by trypsin-Giemsa and the results of chromosome analysis are presented according to ISCN 2020.
- Cytogenetic preparations were hybridized with fluorophore-labeled probes. The following FISH probes were used: the Vysis LSI IGH break-apart (BA) probe, BCL6 BA probe, MYC BA probe, and XL BCL3 BA probe. FISH results were analyzed using fluorescence microscopes equipped with DAPI, fluorescein isothiocyanate (FITC), and tetramethylrhodamine B isothiocyanate (TRITC) fluorescence filters as well as a DAPI/FITC/TRITC triple band-pass filter.
- Browse our recommendations
| Product/Service Types | Description |
| Immunohistochemistry (IHC), Immunofluorescence (IF) Service | Creative Bioarray offers a comprehensive IHC service from project design, and marker selection to image completion and data analysis. We are dedicated to satisfying every customer and assisting them to achieve their specific research goals. |
| Karyotyping (G-Banded) Service | Creative Bioarray has performed many karyotyping (banded chromosome analysis) services for cell line authentication and some specific research. In providing karyotyping information, we can promote the advancement of biology projects. |
| Fluorescent In Situ Hybridization (FISH) Service | Creative Bioarray offers a range of different FISH services, including metaphase and interphase FISH (chromosomal assignment and clone ordering), fibre-FISH (chromosome painting), RNA-FISH (cell-based gene expression assay), M-FISH (multicolor karyotyping), 3D-FISH (on three-dimensionally preserved nuclei), flow-FISH (quantify the length of telomeres), immunoFISH (combined FISH and IHC). |
| Probe | Creative Bioarray provides the most comprehensive list of FISH probes for rapid identification of a wide range of chromosomal aberrations across the genome. |
RESULTS
- G-banding of metaphase spreads obtained from PB, BM, or LN revealed t(14;19)(q32;q13), resulting in the characteristic 14q+ and 19q− morphology, in all 5 cases. (Fig.1, 2, and 3). In case 5, both diploid- and tetraploid-range karyotypes had a single copy of t(14;19)(q32;q13) (Fig. 3). The translocation was confirmed by FISH in all cases, demonstrating colocalization of red-colored centromeric 3' IGH and green-colored telomeric 3' BCL3 probes at q32 of der(14)t(14;19), corresponding to the IGH::BCL3 fusion gene, and red-colored centromeric 5' BCL3 and green-colored telomeric 5' IGH probes at q13 of der(19)t(14;19), corresponding to the reciprocal gene fusion (Fig. 1, 2, and 3). Other 14q32/IGH translocations included t(2;14)(p13;q32) in case 1, t(8;14)(q24;q32) in case 3, and t(8;11;14)(q24;q11;q32) in case 4 (Fig. 1, 2, and 3). In the latter 2 translocations, FISH revealed colocalization of red-colored centromeric 3' IGH and green-colored 3' MYC at q32 of der(14)t(8;14) and der(14)t(8;11;14), respectively, proving the generation of the IGH::MYC fusion gene (Fig. 2). Additional abnormalities recognized by G-banding and FISH were trisomy 12 in cases 1, 2, and 3, 3 copies of MYC in cases 1 and 3, and 4 copies of BCL6 in association with deletion of a red-colored telomeric 5' BCL6 probe in case 4 (Fig. 1 and 2). A CLL FISH panel test to detect del(6q), del(11q), del(13q), and del(17p) was negative in all cases tested (Fig. 1B).
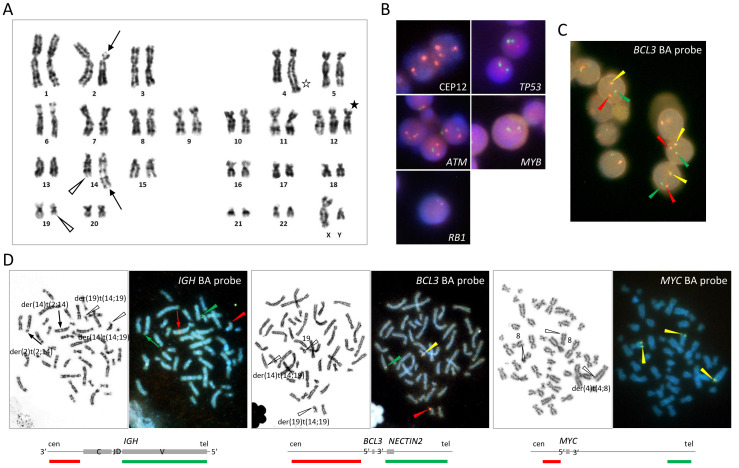 Fig. 1 Cytogenetic findings in case 1.
Fig. 1 Cytogenetic findings in case 1.
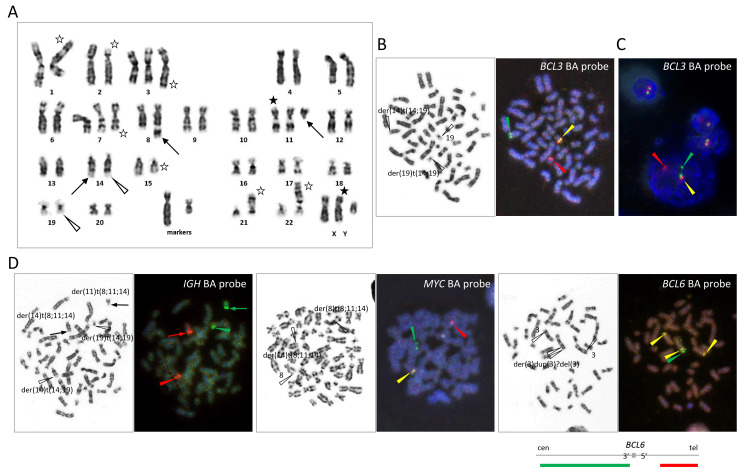 Fig. 2 Cytogenetic findings in case 4.
Fig. 2 Cytogenetic findings in case 4.
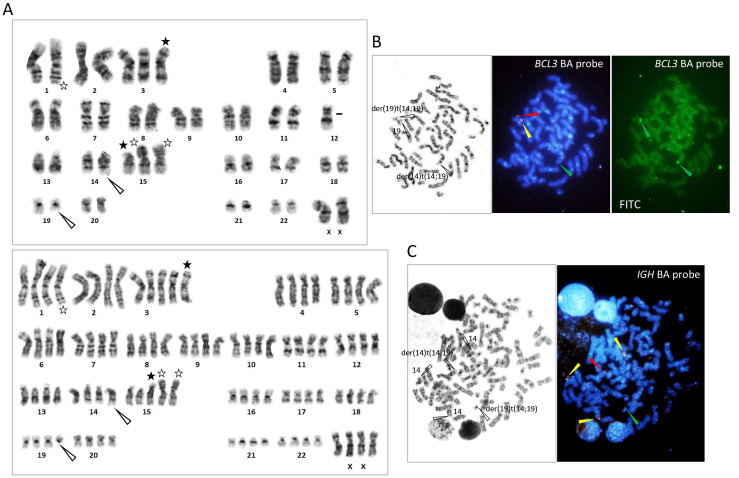 Fig. 3 Cytogenetic findings in case 5.
Fig. 3 Cytogenetic findings in case 5.
- Histopathological examination of the LN biopsy in case 2 revealed diffuse proliferation of small cells admixed with larger prolymphocytes/para immunoblasts. The cells were positive for CD20 and BCL2 and negative for CD5 by IHC. The Ki67 index was 20%. In cases 3 and 4, on the other hand, tumor cells were large and had round, vesicular nuclei with prominent nucleoli; the proportion of tumor cells was lower in the latter case compared with the former (Fig. 4). The cells were negative for CD10 and positive for CD20, MUM1, and BCL2. A fraction of large cells was positive for CD5 in case 3. BCL6 was negative in case 3 and positive in case 4. Thus, cases 3 and 4 were classified into the non-germinal center B-like (non-GCB) category of diffuse large B-cell lymphoma (DLBCL). The tumor cell nuclei of both cases were stained positive by anti-MYC and anti-BCL3 antibodies (Fig. 4). The Ki67 index was >90% in case 3 and 40% in case 4. In case 5, the LN structure was effaced with infiltration of CD3-positive small to medium-sized cells, CD20-positive large B cells, and CD30-positive cells in the setting of proliferation of high-endothelial venules and polymorphic cellular infiltrates composed of histiocytes, plasma cells, and eosinophils (Fig. 5). Some T cells were positive for PD-1 and clusters of BCL6-positive cells were found. CD10 was negative. EBER ISH revealed many EBV-positive cells, most likely corresponding to large B cells (Fig. 5).
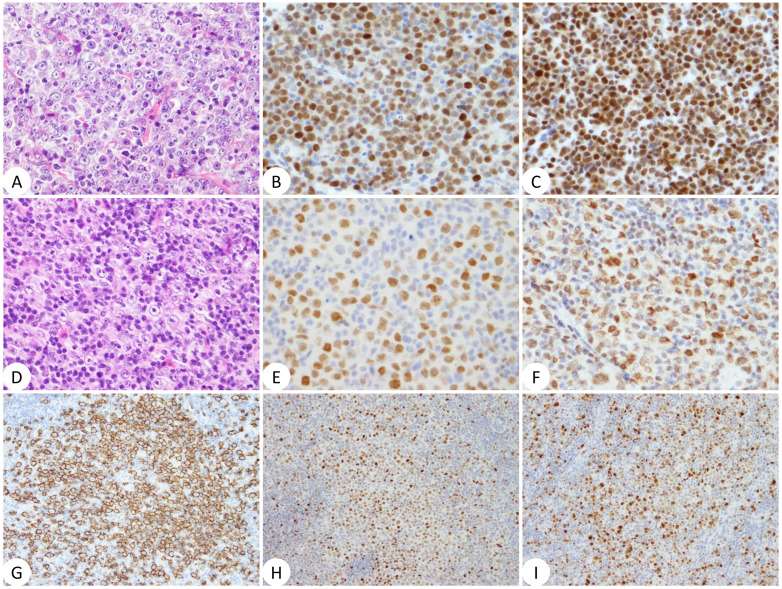 Fig. 4 Histopathology of cases 3 (A to C) and 4 (D to I).
Fig. 4 Histopathology of cases 3 (A to C) and 4 (D to I).
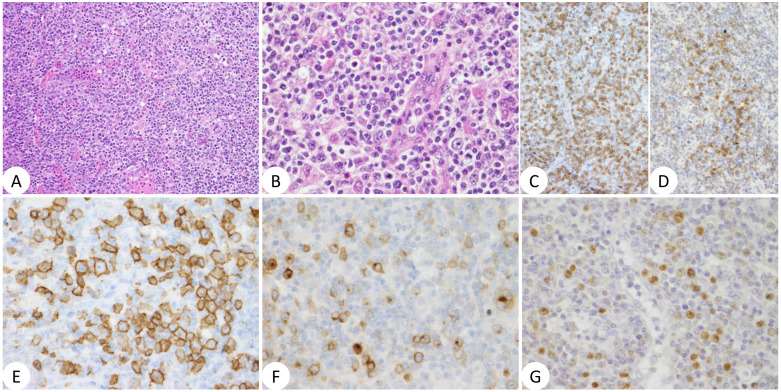 Fig. 5 Histopathology of case 5.
Fig. 5 Histopathology of case 5.
SUMMARY
We characterized 5 B-cell tumors carrying t(14;19)(q32;q13) that creates the IGH::BCL3 fusion gene. The presence of t(14;19)(q32;q13) was confirmed by FISH, showing colocalization of 3' IGH and 3' BCL3 probes on der(14)t(14;19) and 5' BCL3 and 5' IGHprobes on der(19)t(14;19).
RELATED PRODUCTS & SERVICES
Reference
- Ohno H, et al. (2024). "Diverse B-cell tumors associated with t(14;19)(q32;q13)/IGH::BCL3 identified by G-banding and fluorescence in situ hybridization." J Clin Exp Hematop. 64 (1): 21-31.