Featured Products
Our Promise to You
Guaranteed product quality, expert customer support

Uterine/Cervix Tumor Cells
- Product List
- Background
- Applications
- Scientific Data
Cancers of the uterus and cervix remain significant health challenges for women worldwide. The intricate cellular and molecular processes that underlie the development of uterine and cervical tumors have been the focus of intense research. Understanding the biology of uterine and cervical tumor cells is essential for advancing treatment options and improving patient outcomes. This includes investigating the genetic and molecular alterations that drive tumorigenesis, the characteristics of tumor microenvironments, and the mechanisms of metastasis.
Cellular and Molecular Biology of Uterine/Cervix Tumor Cells
- Tumor cell characteristics. Tumor cells in the uterus and cervix exhibit distinct characteristics that differentiate them from normal cells. These include uncontrolled proliferation, evasion of apoptosis, and the ability to invade surrounding tissues. Uterine tumors, such as endometrial carcinoma, often arise from the endometrial lining, while cervical tumors frequently originate from the epithelial cells of the cervix. The cellular architecture of these tumors can vary significantly, with histological subtypes influencing prognosis and treatment decisions.
- Genetic alterations. Genetic mutations play a pivotal role in the development and progression of uterine and cervical tumors. In cervical cancer, the integration of high-risk Human Papillomavirus (HPV) types, particularly HPV 16 and 18, is a well-established factor. These viral oncogenes produce proteins (E6 and E7) that interfere with tumor suppressor functions of p53 and Rb, respectively, leading to cellular transformation. In endometrial cancer, common mutations include alterations in the PTEN, PIK3CA, and KRAS genes, which contribute to the dysregulation of the PI3K/AKT pathway, promoting cell survival and proliferation.
- Metastasis and invasion. The ability of tumor cells to metastasize is a defining feature of malignant tumors. In cervical cancer, metastasis often occurs through lymphatic spread, with the pelvic lymph nodes being the most common sites. Endometrial cancer can also metastasize via lymphatic channels, as well as through hematogenous routes to distant organs. Understanding the molecular pathways that govern these processes, such as epithelial-to-mesenchymal transition (EMT), is essential for developing targeted therapies.
Research and Drug Development
Uterine and cervical tumor cells provide valuable models for cancer research and drug development. Cell lines derived from these tumors, such as HeLa cells (from cervical cancer), have been instrumental in elucidating fundamental biological processes and testing new therapeutic agents. For instance, the study of HeLa cells has led to significant advances in understanding cancer biology and has facilitated the development of targeted therapies that inhibit specific signaling pathways.
Biomarker Discovery
Identifying biomarkers associated with uterine and cervical tumors is critical for early diagnosis and treatment stratification. Recent advancements in genomics and proteomics have enabled the discovery of potential biomarkers that can predict disease progression and response to therapy. For instance, the overexpression of p16INK4a is a well-established marker for HPV-related cervical cancer, aiding in diagnostic accuracy. Moreover, circulating tumor DNA (ctDNA) analysis is emerging as a non-invasive method for monitoring treatment response and detecting recurrence.
Therapeutic Innovations
Innovative therapeutic strategies are being developed based on the cellular and molecular insights gained from studies on uterine and cervical tumor cells. Immunotherapy, for instance, has shown promise in treating advanced cervical cancer, particularly with the use of immune checkpoint inhibitors such as pembrolizumab. Furthermore, targeted therapies that focus on specific genetic alterations, such as PI3K inhibitors for endometrial cancer with PIK3CA mutations, are being explored in clinical trials.
GDF15 Promoted Cervical Cancer Cell Proliferation and Tumor Formation
Growth differentiation factor 15 (GDF15, also known as MIC-1, NAG-1, PTGF-β, and PDF) belongs to the transforming growth factor beta (TGF-β) superfamily. Evidence shows that GDF15 plays an important role in carcinogenesis-related activities, including proliferation, migration, invasion, and angiogenesis in various types of tumors. To further investigate the function of GDF15 in human cervical cancer cells, commercial recombinant human GDF15 (rhGDF15) and human serum albumin (HSA) were added to the media of HeLa and SiHa. An MTT assay revealed that rhGDF15 stimulated the proliferation of HeLa and SiHa cells in a dose-dependent manner. Additionally, cell growth curves and an MTT assay were applied to evaluate the proliferation and viability of the HeLa and SiHa cell lines, which were incubated with PBS, HSA, or rhGDF15. As shown in Fig. 1a and b, HeLa and SiHa cells treated with rhGDF15 grew more rapidly than their respective controls (HeLa PBS and SiHa PBS, HeLa-HSA and SiHa-HSA, P < 0.05). Moreover, the viability of HeLa and SiHa cells treated with rhGDF15 was much higher than that of their respective controls (Fig.1c and d, P < 0.05). Furthermore, the supernatants of the GFP and GDF15-transfected HeLa and SiHa cell clones (GDF15 concentration in the supernatant of HeLa-GFP and SiHa-GFP is 3.64 ± 0.12 ng/ml and 0.98 ± 0.07 ng/ml; GDF15 concentration in the supernatant of HeLa-GDF15 and SiHa-GDF15 is 38.7 ± 0.83 ng/ml and 19.4 ± 0.51 ng/ml respectively, determined by ELISA assay) were collected and incubated with HeLa-GFP and SiHa-GFP cells for 24 h, 48 h, 72 h and 96 h. An MTT study showed that a 50% conditional medium significantly increased the proliferation of HeLa-GFP and SiHa-GFP cells after treatment for 96 h. Conversely, GDF15 knockdown in HT-3 cells using both CRISPR-CAS9 mediated gene editing and shRNA technologies resulted in significant decreases in cell growth and viability (Fig. 1e to h, P < 0.05). All of these results demonstrated that the GDF15 protein stimulated the proliferation of cervical cancer cells in vitro in both an autocrine and a paracrine manner.
To further determine the effects of GDF15 on tumor formation ability in vivo, 6 adult female nude mice in each group were subcutaneously transplanted with HeLa-GDF15 or SiHa-GDF15 and their control cells (Fig. 1m and n). The xenograft assay in BALB/c-nude mice showed that GDF15-overexpressing cells formed much larger tumors in terms of volume (Fig. 1i and j, P < 0.05) and weight (Fig. 1k and l, P < 0.05) than did control cells (HeLa-GFP and SiHa-GFP). Furthermore, IHC revealed that the xenograft tumor tissues formed by HeLa-GDF15 and SiHa-GDF15 cells demonstrated much stronger Ki67 staining scores than those formed by control cells (HeLa-GFP and SiHa-GFP) (Fig. 1o-r, P < 0.05). All of these data suggested that GDF15 promoted tumor formation of cervical cancer cells, potentially via enhanced cell proliferation in vivo.
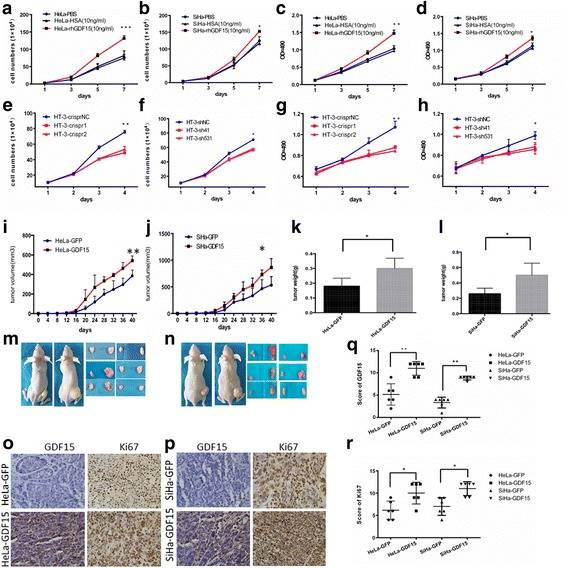 Fig. 1 The proliferation and viability of HeLa and SiHa cells incubated with PBS, HSA(10 ng/ml), or rhGDF15(10 ng/ml) were detected by growth curves (a and b, respectively) and the MTT assay (c and d, respectively). The proliferation and viability of HT-3 cells modified with CRISPR and shRNA were detected by growth curves (e and f, respectively) and the MTT assay (g and h, respectively). Tumor growth curves (i and j, respectively) and tumor weights (k and l, respectively) are shown for HeLa-GDF15 and SiHa-GDF15 cells and control cells (HeLa-GFP and SiHa-GFP). Pictures were of nude mice with tumors from HeLa-GDF15 and SiHa-GDF15 and their control cells (m and n, respectively). Immunohistochemical staining results for Ki67 are shown in tumor xenografts of GDF15-overexpressing HeLa and SiHa cells (o and p, respectively) and the quantitative analysis is shown (q and r, respectively). (Li S, et al, 2018)
Fig. 1 The proliferation and viability of HeLa and SiHa cells incubated with PBS, HSA(10 ng/ml), or rhGDF15(10 ng/ml) were detected by growth curves (a and b, respectively) and the MTT assay (c and d, respectively). The proliferation and viability of HT-3 cells modified with CRISPR and shRNA were detected by growth curves (e and f, respectively) and the MTT assay (g and h, respectively). Tumor growth curves (i and j, respectively) and tumor weights (k and l, respectively) are shown for HeLa-GDF15 and SiHa-GDF15 cells and control cells (HeLa-GFP and SiHa-GFP). Pictures were of nude mice with tumors from HeLa-GDF15 and SiHa-GDF15 and their control cells (m and n, respectively). Immunohistochemical staining results for Ki67 are shown in tumor xenografts of GDF15-overexpressing HeLa and SiHa cells (o and p, respectively) and the quantitative analysis is shown (q and r, respectively). (Li S, et al, 2018)
Enhanced Expression of Mcm Proteins in Uterine/Cervix Tumor Cells
Minichromosome maintenance proteins (Mcm) 2-7 play essential roles in eukaryotic DNA replication. Several reports have indicated the usefulness of Mcm proteins as markers of cancer cells in histopathological diagnosis. Total cellular proteins (2×107 cells·mL-1) and proteins bound to chromatin (4×107 cells·mL-1) were prepared from logarithmically growing HeLa uterine cervical carcinoma cells and human normal fibroblast WI-38 cells. They were analyzed by Western blotting using anti-Mcm2, 3, 4, 5, 6, and 7 Ig (Fig. 2). On comparing the amounts of Mcm in total cellular proteins and chromatin-bound proteins, approximately one-third of total Mcm protein was found to be recovered in the chromatin-bound fraction in HeLa cells. Thus, a considerable portion of Mcm protein is present in the nucleoplasm and/or easily released from chromatin. Titration of these two fractions in the Western blot analysis shows that Mcm2-7 proteins are 5-14 times more abundant in HeLa cells than in WI-38 cells for the total cellular proteins and are 6-13 times for chromatin-bound proteins (Fig. 2 and Table 1). Using purified human Mcm proteins as a standard, it was calculated that approximately 1.5-2.5×106 molecules of Mcm2, 4, 6, and 7 proteins are present in a single HeLa cell on average, and 0.1-0.5×106 molecules in a single WI-38 cell (Table 1).
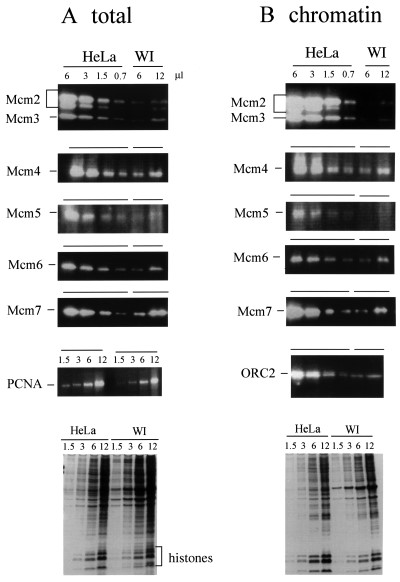 Fig. 2 Mcm proteins in total cell extracts and chromatin-bound fraction from HeLa and WI 38 cells. (A) Different volumes (0.7-12 µL) of total cellular proteins (equivalent to 2×104 cells·µL-1) were separated in SDS-polyacrylamide gel, transferred to a membrane, and analyzed by immunoblotting using anti-Mcm and anti-PCNA Ig. (B) Chromatin-bound proteins (4×104 cells·µL-1) were analyzed by immunoblotting using anti-Mcm and anti-ORC2 Ig. (Ishimi Y, et al., 2003)
Fig. 2 Mcm proteins in total cell extracts and chromatin-bound fraction from HeLa and WI 38 cells. (A) Different volumes (0.7-12 µL) of total cellular proteins (equivalent to 2×104 cells·µL-1) were separated in SDS-polyacrylamide gel, transferred to a membrane, and analyzed by immunoblotting using anti-Mcm and anti-PCNA Ig. (B) Chromatin-bound proteins (4×104 cells·µL-1) were analyzed by immunoblotting using anti-Mcm and anti-ORC2 Ig. (Ishimi Y, et al., 2003)
Table 1. Quantitation and comparison of Mcm amounts. From the data in Fig. 2 and others, the concentrations of Mcm proteins, ORC2, and PCNA were compared between HeLa and WI-38 cells for their two fractions, total cellular protein, and chromatin-bound protein. (Ishimi Y, et al., 2003)
| Numbers of HeLa/WI | Numbers of molecules (total) | |||
| Total | Chromatin | HeLa | WI | |
| Mcm 2 | 14 (13-16) | 12 | 1.6×106 | 1×105 |
| 3 | 10 (8-12) | 11 (7-15) | ||
| 4 | 5 (4-7) | 6 (6-7) | 2.5×106 | 5.2×105 |
| 5 | 7 | ND | ||
| 6 | 10 (5-15) | 10 (4-14) | 2×106 | 1.8×105 |
| 7 | 9 (6-10) | 13 (10-16) | 1.5×106 | 2.1×105 |
| ORC2 | 8 | |||
| PCNA | 1.5 (1.4-1.9) | |||
| Histones | 1.5-2 | |||
Description: Melanin producing. Originated from the same site of HMV-I.
Description: Large cell non-keratinizing squamouse carcinoma. Stage Ib. HPV type 18 integrated.
Description: Derived from a single human as TCO-1. Said CEA, CA125, TPA (+).
Description: Poorly differentiated adenocarcinoma. Cell growth is slow.
Description: Glassy cell carcinoma. TA-4, CA125, neuron-specific enolase producing.
Description: Keratinizing cervix squamous carcinoma, CEA(+).
Description: Integrating HPV16, neurosecretory granules(+)
Description: Suspension culturable. Useful for JIS medium inspection
Description: Human uterus squamous cell carcinoma cell line.
Description: Human uterus endometrioid adenocarcinoma cell line.

