- You are here: Home
- Resources
- Explore & Learn
- Exosome
- Techniques for Exosome Quantification
Support
-
Cell Services
- Cell Line Authentication
- Cell Surface Marker Validation Service
-
Cell Line Testing and Assays
- Toxicology Assay
- Drug-Resistant Cell Models
- Cell Viability Assays
- Cell Proliferation Assays
- Cell Migration Assays
- Soft Agar Colony Formation Assay Service
- SRB Assay
- Cell Apoptosis Assays
- Cell Cycle Assays
- Cell Angiogenesis Assays
- DNA/RNA Extraction
- Custom Cell & Tissue Lysate Service
- Cellular Phosphorylation Assays
- Stability Testing
- Sterility Testing
- Endotoxin Detection and Removal
- Phagocytosis Assays
- Cell-Based Screening and Profiling Services
- 3D-Based Services
- Custom Cell Services
- Cell-based LNP Evaluation
-
Stem Cell Research
- iPSC Generation
- iPSC Characterization
-
iPSC Differentiation
- Neural Stem Cells Differentiation Service from iPSC
- Astrocyte Differentiation Service from iPSC
- Retinal Pigment Epithelium (RPE) Differentiation Service from iPSC
- Cardiomyocyte Differentiation Service from iPSC
- T Cell, NK Cell Differentiation Service from iPSC
- Hepatocyte Differentiation Service from iPSC
- Beta Cell Differentiation Service from iPSC
- Brain Organoid Differentiation Service from iPSC
- Cardiac Organoid Differentiation Service from iPSC
- Kidney Organoid Differentiation Service from iPSC
- GABAnergic Neuron Differentiation Service from iPSC
- Undifferentiated iPSC Detection
- iPSC Gene Editing
- iPSC Expanding Service
- MSC Services
- Stem Cell Assay Development and Screening
- Cell Immortalization
-
ISH/FISH Services
- In Situ Hybridization (ISH) & RNAscope Service
- Fluorescent In Situ Hybridization
- FISH Probe Design, Synthesis and Testing Service
-
FISH Applications
- Multicolor FISH (M-FISH) Analysis
- Chromosome Analysis of ES and iPS Cells
- RNA FISH in Plant Service
- Mouse Model and PDX Analysis (FISH)
- Cell Transplantation Analysis (FISH)
- In Situ Detection of CAR-T Cells & Oncolytic Viruses
- CAR-T/CAR-NK Target Assessment Service (ISH)
- ImmunoFISH Analysis (FISH+IHC)
- Splice Variant Analysis (FISH)
- Telomere Length Analysis (Q-FISH)
- Telomere Length Analysis (qPCR assay)
- FISH Analysis of Microorganisms
- Neoplasms FISH Analysis
- CARD-FISH for Environmental Microorganisms (FISH)
- FISH Quality Control Services
- QuantiGene Plex Assay
- Circulating Tumor Cell (CTC) FISH
- mtRNA Analysis (FISH)
- In Situ Detection of Chemokines/Cytokines
- In Situ Detection of Virus
- Transgene Mapping (FISH)
- Transgene Mapping (Locus Amplification & Sequencing)
- Stable Cell Line Genetic Stability Testing
- Genetic Stability Testing (Locus Amplification & Sequencing + ddPCR)
- Clonality Analysis Service (FISH)
- Karyotyping (G-banded) Service
- Animal Chromosome Analysis (G-banded) Service
- I-FISH Service
- AAV Biodistribution Analysis (RNA ISH)
- Molecular Karyotyping (aCGH)
- Droplet Digital PCR (ddPCR) Service
- Digital ISH Image Quantification and Statistical Analysis
- SCE (Sister Chromatid Exchange) Analysis
- Biosample Services
- Histology Services
- Exosome Research Services
- In Vitro DMPK Services
-
In Vivo DMPK Services
- Pharmacokinetic and Toxicokinetic
- PK/PD Biomarker Analysis
- Bioavailability and Bioequivalence
- Bioanalytical Package
- Metabolite Profiling and Identification
- In Vivo Toxicity Study
- Mass Balance, Excretion and Expired Air Collection
- Administration Routes and Biofluid Sampling
- Quantitative Tissue Distribution
- Target Tissue Exposure
- In Vivo Blood-Brain-Barrier Assay
- Drug Toxicity Services
Techniques for Exosome Quantification
Exosomes are small extracellular vesicles that play crucial roles in intercellular communication and contribute to various physiological and pathological processes. As the field of exosome research continues to expand, the need for reliable and accurate techniques for exosome quantification becomes increasingly important. Exosome quantification mainly depends on their characteristic physical properties, such as size, mass, and density, or membrane proteins present on their surface.
Nano-Sight Nanoparticle Tracking Analysis (NTA)
Nano-sight NTA is a widely employed process for quantifying exosomes. Technical abbreviations will be explained upon first usage. It exploits the principles of light scattering and Brownian motion to observe and monitor individual exosomes in liquid. Videos of the particles' Brownian motion are captured by the device, and the software tracks and evaluates the particle trajectories for size and concentration determination.
NTA presents high-resolution size distribution profiles and concentration measurements of exosomes. It allows researchers to monitor changes in exosome populations under different conditions or during disease progression. The quantification process occurs in real time, allowing dynamic monitoring of exosome release and uptake by cells, thereby facilitating comprehensive exosome analysis.
Tunable Resistive Pulse Sensing (TRPS)
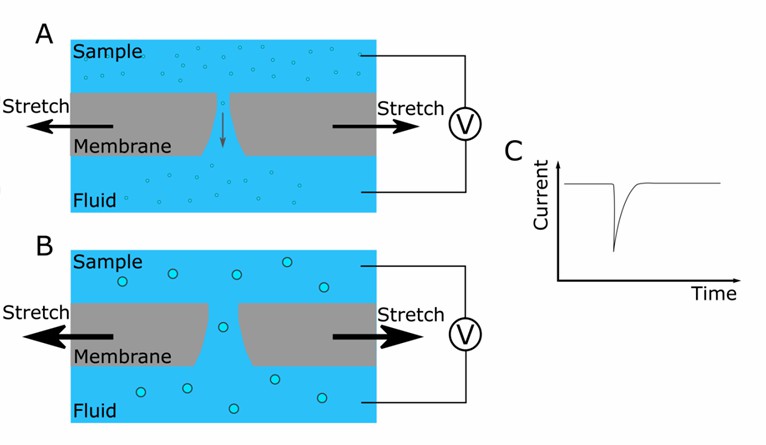 Fig. 1 Schematic representation of the instrument. (Koritzinsky EH, et al., 2017)
Fig. 1 Schematic representation of the instrument. (Koritzinsky EH, et al., 2017)
TRPS measures changes in electrical resistance as individual exosomes travel through a nanopore. The magnitude and duration of these resistance changes determine the size and concentration of exosomes in the sample.
TRPS offers high sensitivity and resolution in exosome quantification. It detects and sizes exosomes within a broad size range, providing valuable insights into the heterogeneity of exosome populations. TRPS is capable of analyzing various types of nanoparticles, aiding in the distinction of exosomes from other vesicles or impurities in the sample. Additionally, TRPS holds potential for the analysis of other nanoparticle species.
Surface Plasmon Resonance (SPR)
SPR is a label-free technique commonly used for studying biomolecular interactions. It can also be adapted for exosome quantification by immobilizing exosome-specific ligands on a sensor chip. When exosomes bind to the ligands on the chip surface, changes in the refractive index near the surface lead to a shift in the resonance angle, which can be measured and correlated with exosome concentration.
SPR offers real-time monitoring of exosome binding events and provides quantitative information on exosome kinetics and affinity. It is a versatile technique that can be used to study exosome interactions with other molecules, such as proteins or nucleic acids, providing insights into exosome-mediated signaling pathways.
Electron Microscopy (EM)
EM is a high-resolution imaging technique that allows for the direct visualization of exosomes. It involves the use of an electron microscope to image exosomes in biological samples. In transmission electron microscopy (TEM), exosomes are visualized by staining and embedding samples in resin, followed by ultra-thin sectioning and imaging under an electron beam. Scanning electron microscopy (SEM) provides surface morphology information of exosomes.
EM provides detailed structural information about exosomes, including their size, shape, and internal cargo. While it does not provide quantitative data on exosome concentration, EM is often used in conjunction with other quantification techniques to validate the presence and morphology of exosomes.
Creative Bioarray Relevant Recommendations
Creative Bioarray offers a range of options to meet most exosome quantitation demands, including ELISA-based immunoaffinity capture (IAC) assay, NTA, asymmetrical-flow field flow fractionation (AF4) coupled with multi-detection assay, dynamic light scattering (DLS) assay, and SPR assay.
Reference
- Koritzinsky EH, et al. (2017). "Quantification of Exosomes." J Cell Physiol. 232 (7), 1587-1590.
For research use only. Not for any other purpose.

