- You are here: Home
- Resources
- Explore & Learn
- Toxicokinetics & Pharmacokinetics
- Predictive Modeling of Metabolic Drug Toxicity
Support
-
Cell Services
- Cell Line Authentication
- Cell Surface Marker Validation Service
-
Cell Line Testing and Assays
- Toxicology Assay
- Drug-Resistant Cell Models
- Cell Viability Assays
- Cell Proliferation Assays
- Cell Migration Assays
- Soft Agar Colony Formation Assay Service
- SRB Assay
- Cell Apoptosis Assays
- Cell Cycle Assays
- Cell Angiogenesis Assays
- DNA/RNA Extraction
- Custom Cell & Tissue Lysate Service
- Cellular Phosphorylation Assays
- Stability Testing
- Sterility Testing
- Endotoxin Detection and Removal
- Phagocytosis Assays
- Cell-Based Screening and Profiling Services
- 3D-Based Services
- Custom Cell Services
- Cell-based LNP Evaluation
-
Stem Cell Research
- iPSC Generation
- iPSC Characterization
-
iPSC Differentiation
- Neural Stem Cells Differentiation Service from iPSC
- Astrocyte Differentiation Service from iPSC
- Retinal Pigment Epithelium (RPE) Differentiation Service from iPSC
- Cardiomyocyte Differentiation Service from iPSC
- T Cell, NK Cell Differentiation Service from iPSC
- Hepatocyte Differentiation Service from iPSC
- Beta Cell Differentiation Service from iPSC
- Brain Organoid Differentiation Service from iPSC
- Cardiac Organoid Differentiation Service from iPSC
- Kidney Organoid Differentiation Service from iPSC
- GABAnergic Neuron Differentiation Service from iPSC
- Undifferentiated iPSC Detection
- iPSC Gene Editing
- iPSC Expanding Service
- MSC Services
- Stem Cell Assay Development and Screening
- Cell Immortalization
-
ISH/FISH Services
- In Situ Hybridization (ISH) & RNAscope Service
- Fluorescent In Situ Hybridization
- FISH Probe Design, Synthesis and Testing Service
-
FISH Applications
- Multicolor FISH (M-FISH) Analysis
- Chromosome Analysis of ES and iPS Cells
- RNA FISH in Plant Service
- Mouse Model and PDX Analysis (FISH)
- Cell Transplantation Analysis (FISH)
- In Situ Detection of CAR-T Cells & Oncolytic Viruses
- CAR-T/CAR-NK Target Assessment Service (ISH)
- ImmunoFISH Analysis (FISH+IHC)
- Splice Variant Analysis (FISH)
- Telomere Length Analysis (Q-FISH)
- Telomere Length Analysis (qPCR assay)
- FISH Analysis of Microorganisms
- Neoplasms FISH Analysis
- CARD-FISH for Environmental Microorganisms (FISH)
- FISH Quality Control Services
- QuantiGene Plex Assay
- Circulating Tumor Cell (CTC) FISH
- mtRNA Analysis (FISH)
- In Situ Detection of Chemokines/Cytokines
- In Situ Detection of Virus
- Transgene Mapping (FISH)
- Transgene Mapping (Locus Amplification & Sequencing)
- Stable Cell Line Genetic Stability Testing
- Genetic Stability Testing (Locus Amplification & Sequencing + ddPCR)
- Clonality Analysis Service (FISH)
- Karyotyping (G-banded) Service
- Animal Chromosome Analysis (G-banded) Service
- I-FISH Service
- AAV Biodistribution Analysis (RNA ISH)
- Molecular Karyotyping (aCGH)
- Droplet Digital PCR (ddPCR) Service
- Digital ISH Image Quantification and Statistical Analysis
- SCE (Sister Chromatid Exchange) Analysis
- Biosample Services
- Histology Services
- Exosome Research Services
- In Vitro DMPK Services
-
In Vivo DMPK Services
- Pharmacokinetic and Toxicokinetic
- PK/PD Biomarker Analysis
- Bioavailability and Bioequivalence
- Bioanalytical Package
- Metabolite Profiling and Identification
- In Vivo Toxicity Study
- Mass Balance, Excretion and Expired Air Collection
- Administration Routes and Biofluid Sampling
- Quantitative Tissue Distribution
- Target Tissue Exposure
- In Vivo Blood-Brain-Barrier Assay
- Drug Toxicity Services
Predictive Modeling of Metabolic Drug Toxicity
Acute toxicity refers to the toxic effect or death of the body due to a single exposure or multiple exposures to exogenous chemicals within 24 hours, which is an important indicator of drug safety evaluation. Predicting the potential toxicity of new drug candidates is a critical step in the drug discovery and development process. Determining the safety profile of drugs is essential to ensure patient well-being and regulatory compliance. In recent years, predictive modeling approaches have emerged as valuable tools for assessing the toxicity of drugs. These models utilize computational algorithms and large datasets to predict various types of drug toxicity, including acute oral toxicity, mutagenicity, carcinogenicity, hERG toxicity, hepatotoxicity, and endocrine disruption.
Prediction of Acute Oral Toxicity
Acute toxicity of drugs can be categorized into oral, percutaneous, and inhalation acute toxicity. Since drugs are mainly taken orally, the prediction model constructed in this way is the key research object in the R&D process. Existing prediction models include the local model and the global model.
Prediction of Mutagenicity
Mutagenicity is the alteration of DNA in the cell nucleus caused by a compound. Since such changes are passed on as the cell divides, the mutagenicity of a compound is an important indicator of a drug's safety. Bruce Ames proposed a mutant testing method (Ames test) in 1983 and detected 175 known carcinogens. The Ames test is commonly used as a pre-test method and is widely used as a model system for the toxicity and mutagenicity safety assessment of chemicals.
Carcinogenicity Prediction
Carcinogenicity is one of the most important indicators for drug safety evaluation. Predictive models for the carcinogenicity of compounds can also be categorized into local and global models. Local models mainly focus on congeners such as N-nitroso compounds, aromatic amine compounds, and polycyclic aromatic compounds.
Prediction of hERG Toxicity
hERG (human Ether-à-go-go-Related Gene) toxicity refers to the potential of a drug to inhibit the hERG potassium ion channels in the heart, which can lead to life-threatening cardiac arrhythmias. Predictive modeling techniques integrate chemical descriptors, structural information, and hERG inhibition data to develop models capable of predicting hERG toxicity. These models aid in the identification of compounds with a high risk of hERG inhibition, enabling researchers to prioritize drug candidates with a lower risk of cardiac side effects.
Prediction of Drug Hepatotoxicity
Hepatotoxicity caused by drug-induced liver injury (DILI) is a common adverse drug reaction. DILI not only jeopardizes the health of drug users, but also is the main reason for drug development failure, restriction of use, and withdrawal of drugs from the market.
With the development of modern life sciences, computers, bioinformatics, and other technologies, computational models based on the molecular structure of drugs have been widely applied to the prediction of drug-induced hepatotoxicity. By using Bayesian methods, deep learning techniques, and substructure pattern recognition methods. These computational models have obtained good results in the practical application of predicting hepatotoxicity.
Prediction of Drug Endocrine Disruption
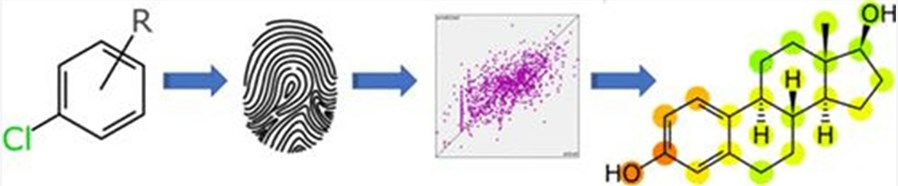 Fig. 1 Machine learning models for endocrine disruption prediction. (Zorn KM, et al., 2020)
Fig. 1 Machine learning models for endocrine disruption prediction. (Zorn KM, et al., 2020)
Endocrine-disrupting chemicals (EDCs) are compounds in the environment that are capable of disrupting the human endocrine system, which in turn can have adverse effects on the reproductive, developmental, neurological, and immune systems. Earlier, predictive models for EDCs were mostly built based on statistical algorithms and molecular descriptors, and most of them required limited in vitro data support. Recently, machine learning methods have emerged as a way to predict molecular structures prospectively, providing a more comprehensive and reliable prediction of the endocrine-disrupting properties of compounds with lower computational costs.
Creative Bioarray Relevant Recommendations
Creative Bioarray specializes in identifying potential drug toxicity to different tissues and organs using a panel of in vitro cell-based assays and is an ideal partner for testing the potential toxicities of candidate drug compounds.
Reference
- Zorn KM, et al. (2020). "Machine Learning Models for Estrogen Receptor Bioactivity and Endocrine Disruption Prediction." Environ Sci Technol. 54 (19), 12202-12213.
For research use only. Not for any other purpose.

