Featured Products
Our Promise to You
Guaranteed product quality, expert customer support

ONLINE INQUIRY
- Specification
- Background
- Scientific Data
- Q & A
- Customer Review
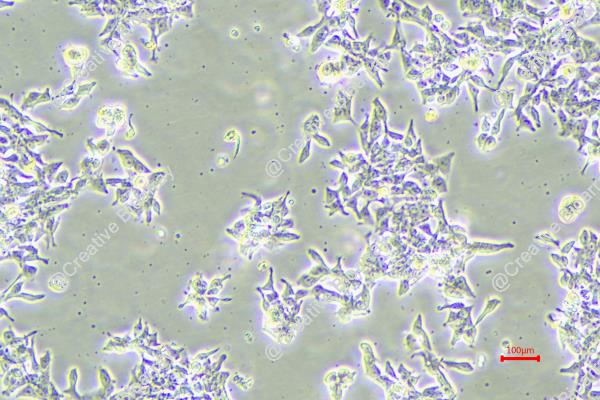
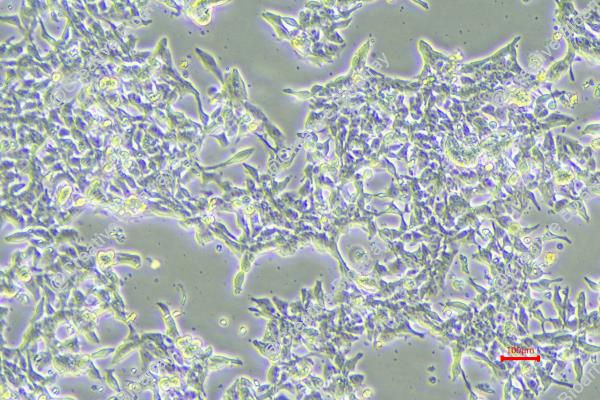
The GP2D cell line was established from an adenocarcinoma identified as a local recurrence of Duke's grade B colon cancer. This classification indicates that the tumor was originally diagnosed at a stage where there was significant local invasion but no distant metastasis. The background of GP2D is particularly noteworthy as it originated from a 71-year-old female, reflecting a demographic commonly affected by colorectal cancer, where age and sex can play crucial roles in tumor characteristics and patient outcomes.
The GP2D cell line is categorized as a poorly differentiated carcinoma, which suggests that the malignant cells exhibit a high degree of abnormality compared to normal colon tissue. Poorly differentiated tumors often demonstrate aggressive behavior, increased propensity for metastasis, and poorer prognosis when compared to well-differentiated tumors. This makes GP2D a valuable model for studying aggressive colorectal cancers, as it can help researchers explore the underlying mechanisms driving tumor progression and resistance to treatment.
Furthermore, the establishment of GP2D has been validated through short tandem repeat (STR) profiling, which confirms its genetic identity and origin from the same adenocarcinoma as the related GP5D cell line. This genetic verification ensures that studies conducted with GP2D can yield reliable and reproducible results when investigating drug responses or molecular pathways associated with poorly differentiated CRC. The insights obtained from GP2D are crucial for the development of therapeutic strategies aimed at treating patients with similar aggressive tumor profiles.
Genetic Analyses of GP2d and GP5d Cell Lines
Two morphologically distinct cell lines, GP2d and GP5d, derived from the same adenocarcinoma of the colon, have been established and characterized. Genetic analyses were performed to determine whether the differing morphologies of the 2 clones arose as a consequence of a detectable genetic difference between the cells. Confirmation of the genetic identity of the cells was obtained by DNA fingerprinting. DNA profiles of GP2d and GP5d were shown to match for each of the 4 single locus probes used; profiles of control DNA obtained from HT29 cells showed no match with either GP2d or GP5d.
Metaphase spreads examined from both cell lines showed identical female karyotypes (Fig. 1). Each possessed 46 chromosomes and both had an interstitial deletion from the long arm of chromosome 5 in the region 14q to 22q. This deletion is typical of colonic adenocarcinomas. Both clones also possessed additional material on the long arm of chromosome 10; this appeared to be an inverted duplication of bands 1Oq11 and 10q21. This region of chromosome 10 is known to contain the ret proto-oncogene.
Direct sequencing of the Ki-ras gene detected a G to A transition in codon 12 in both cell lines, resulting in a Gly to Asp substitution (Fig. 2). Both cell lines contained a normal copy of the Ki-ras gene, as indicated by the equal intensity of the bands derived from the normal and mutant genes. p53 mutations were examined by SSCP analysis; no band shifts were evident in either cell line compared with normal DNA amplification products for exons 5-8 of the p53 gene.
Since karyotype analysis indicated an abnormality on chromosome 10 in the region of the ret proto-oncogene, the 2 colon carcinoma cell lines were analyzed for rearrangement of this gene by Southern hybridization with a ret cDNA probe which detects fragments of 6.3 kb and 6.6 kb after EcoRI digestion (Fig. 3). Rearrangement of the ret gene results in the juxtaposition of the ret transmembrane and tyrosine kinase domains with other 5' expressed sequences. This rearrangement occurs within an intronic sequence contained in the 6.3-kb EcoRI fragment and can be detected by an alteration in the size of this fragment. Neither of the carcinoma cell lines showed any evidence of alteration in this fragment (Fig. 3), and no additional bands were observed even after prolonged exposure to the autoradiogram. Hybridization was also performed on BamHI-digested DNA, but again no rearrangements were detected.
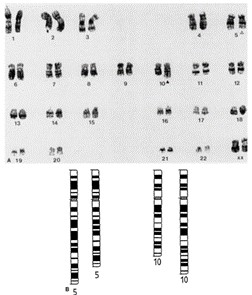 Fig. 1 Karyotypic analysis of GP2d. The figure shows a typical chromosome spread obtained for GP2d which possesses a female kat-yotype of 46 chromosomes (a). The abnormalities are represented diagrammatically in (b). (Solic N, et al., 1995)
Fig. 1 Karyotypic analysis of GP2d. The figure shows a typical chromosome spread obtained for GP2d which possesses a female kat-yotype of 46 chromosomes (a). The abnormalities are represented diagrammatically in (b). (Solic N, et al., 1995)
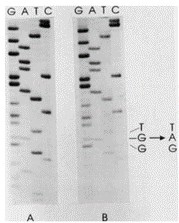 Fig. 2 Analysis of the Ki-ras gene in GP2d and GP5d. The figure shows the autoradiograph of the sequencing gels obtained for GP2d (a) and GP5d (b). (Solic N, et al., 1995)
Fig. 2 Analysis of the Ki-ras gene in GP2d and GP5d. The figure shows the autoradiograph of the sequencing gels obtained for GP2d (a) and GP5d (b). (Solic N, et al., 1995)
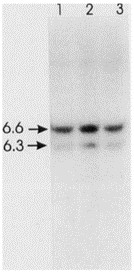 Fig. 3 Analysis of the ret gene in GP2d and GP5d. Southern blot analysis of EcoRI-digested DNA extracted from normal human lymphocytes (lanes 1) and GP2d and GP5d (lanes 2 and 3) and hybridized with a ret cDNA probe. (Solic N, et al., 1995)
Fig. 3 Analysis of the ret gene in GP2d and GP5d. Southern blot analysis of EcoRI-digested DNA extracted from normal human lymphocytes (lanes 1) and GP2d and GP5d (lanes 2 and 3) and hybridized with a ret cDNA probe. (Solic N, et al., 1995)
E-Cadherin in Human Colorectal Carcinoma Cells
Human colorectal cancer cell lines derived from patients were screened for E-cadherin mutations. Single-base deletion mutations were found in nucleotide repeat regions at codons 120 and 126, resulting in frameshifts and premature stops in four RER+ colorectal cancer cell lines (LS174T, HCT116, GP2d, and GP5d) derived from three patients (Fig. 4). These truncations lie in the N-terminal, extracellular domain of the protein and are predicted to generate small, secreted E-cadherin fragments with loss of surface expression and no cellular adhesive capacity. Such small products may even interfere with the normal function of full-length E-cadherin molecules. These frameshift mutations were present as heterozygotes, although an additional single-base substitution was detected at the exon 8 splice donor site in LS174T, with apparent retention of a wild-type allele in HCT116, GP2d, and GP5d.
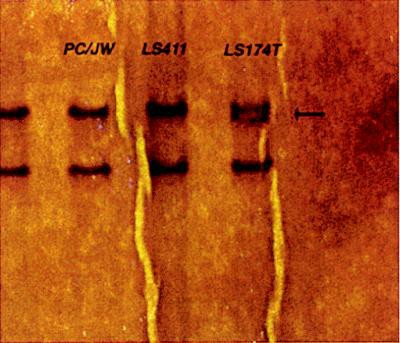 Fig. 4 Single-strand conformational polymorphism (SSCP) analysis of exon 3 of E-cadherin, showing the heterozygous band mobility of LS174T (arrow). (Efstathiou JA, et al., 1999)
Fig. 4 Single-strand conformational polymorphism (SSCP) analysis of exon 3 of E-cadherin, showing the heterozygous band mobility of LS174T (arrow). (Efstathiou JA, et al., 1999)
Biotin, folic acid, niacinamide, pantothenic acid, pyridoxine, riboflavin, thiamine, and vitamin B12 are all common components of culture media.
Ask a Question
Average Rating: 5.0 | 1 Scientist has reviewed this product
Incredible experience
I had an incredible experience with Creative Bioarray's cell culture products. Everything went smoothly and I was very satisfied with the results.
19 June 2022
Ease of use
After sales services
Value for money
Write your own review