DNA Content Analysis
The two most prominent features of the cell cycle are the synthesis and replication of nuclear DNA, and the cell division process itself (mitosis).
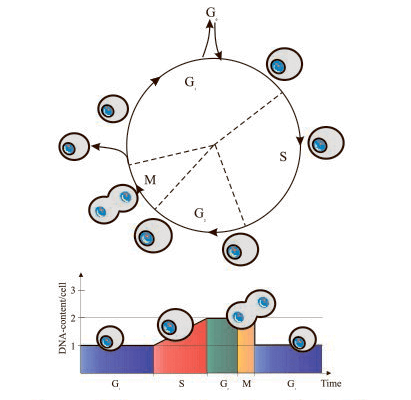
This cell cycle detection method relies on the univariate analysis of cellular DNA content. DNA content can be measured by fluorescent staining which allows the cells to differentiate in G0/G1, S phase, and G2/M, as well as exhibits an emission signal proportional to the amount of DNA. DNA content analysis is typically performed on fixed or permeabilized cells using cell permeable nucleic acid dyes or impermeable nucleic acid stains. The most common DNA binding dyes currently in use are the blue-excited propidium iodide (PI) and the UV-excited diamidino-phenylindole (DAPI) dye. PI is a DNA fluorochrome that binds to DNA or double-stranded RNA, therefore is almost always used with RNAse to remove RNA. While DAPI binds to the small groove of the DNA helix specifically, so it does not require incubation with RNase.
This single time-point measurement reveals the percentage of cells in G0/G1, S, and G2/M, but it does not provide information on cell cycle kinetics.
Common Dyes
Some DNA dyes can’t stain living cells. The samples must be fixed and permeabilized to allow the dyes to enter the cells.
- PI – Blue laser, also stains RNA
- 7-AAD – Helium neon laser
- DAPI – UV laser
Some DNA dyes are membrane permeable and can be used to stain living, intact cells.
- DRAQ5 – Red laser
- Hoechst 33342 – UV laser
Materials and Reagents
Centrifuge
Flow cytometer
70% ethanol
Phosphate buffered saline (PBS)
PI staining solution (0.1% (v/v) Triton X-100, 10 μg/mL PI, and 100 μg/mL RNase A in PBS)
DAPI staining solution (0.1% (v/v) Triton X-100 and 1 μg/mL DAPI in PBS)
PI Staining Procedure
- For suspension cells or hematologic samples, harvest cells by centrifuging 5 min at 500 g. Wash once with PBS. Count cells and thoroughly resuspend 1 x 106 to 2 x 106 cells in 0.5 mL PBS.
- For adherent cells, harvest cells by trypsinization and centrifuge for 5 min at 500 g. Wash once with PBS. Count cells and thoroughly resuspend 1 x 106 to 2 x 106 cells in 0.5 mL PBS.
- Fix cells by transferring this suspension into centrifuge tubes containing 4.5 mL of 70% ethanol, keep on ice for at least 2 h.
Note: 1) It is essential to have a single cell suspension when cells are mixed with ethanol. 2) Cells may be stored in 70% ethanol for weeks at 4°C. - Centrifuge the ethanol-suspended cells for 5 min at 300 g. Decant ethanol thoroughly.
Note: Cell pellet may be loose. Make sure that no cells are lost in this and subsequent washing steps. - Suspend the cell pellet in 5 mL of PBS, wait approximately 30 sec and centrifuge for 5 min at 300 g.
- Resuspend the cell pellet in 1 mL of PI staining solution. Incubate in the dark at room temperature for 30 min, or at 37°C for 10 min.
- Transfer samples to the flow cytometer to measure cell fluorescence. The maximum excitation of PI bound to DNA is at 536 nm, and emission is at 617 nm. Blue (488 nm) or green light lines of lasers are optimal for the excitation of PI fluorescence.
DAPI Staining Procedure
- Suspend the cell pellet in 1 mL of DAPI staining solution. Incubate for 10 min at room temperature in the dark.
- Transfer the sample to the flow cytometer and measure cell fluorescence. The maximum excitation of DAPI bound to DNA is at 359 nm, and emission is at 461 nm. For fluorescence excitation, using the available UV light laser line at the wavelength closest to 359 nm. Cell fluorescence can be measured at wavelengths between 450 and 500 nm by combination of appropriate dichroic mirrors and emission filters.
Tips for Consistent Staining
- Count the cells and calculate the cell concentration
- Use the same number of cells in each sample
- Use sufficient dye
- Stain overnight to achieve equilibrium
- Get a good single cell suspension before fixing
Frequently Asked Questions
Q1: What about RNA?
A1: In addition to DNA, PI also stains RNA, so cells must be treated with RNase. RNase treatment is not required when using DAPI.
Q2: Can cells be alive?
A2: Additional reagents may be required to inhibit the efflux pumps that actively export the dye. Be aware that this staining and subsequent exposure to laser light may result in DNA damage in the cells.
Q3: Which fixative is best?
A3: Alcohols typically result in better staining than crosslinking fixatives. However, the highly fragmented DNA in apoptotic cells may be lost without crosslinking.
Q4: Whole cells or nuclei?
A4: Some protocols suggest lyse the plasma membrane, which both decrease auto-fluorescence and avoid staining of cytoplasmic components. Keep in mind that the nuclear membrane breaks down during mitosis, so lysing the plasma membrane can disperse these chromosomes.
Q5: Is co-staining possible?
A5: One option is to add a membrane permeable DNA dye to existing surface staining protocol. Alternatively, after surface staining, do a gentle crosslinking fixation, permeabilize the cells, and add a DNA dye.
References
- Pozarowski P, et al.; Analysis of cell cycle by flow cytometry. Methods Mol Biol, 2004, 281: 301-311.
- Darzynkiewicz Z. Critical aspects in analysis of cellular DNA content. Current Protocols in Cytometry, 2011, 56: 7.2.1-7.2.8.
- Darzynkiewicz Z, et al.; DNA content measurement for DNA ploidy and cell cycle analysis. Current Protocols in Cytometry, 2001, 00: 7.5.1-7.5.24.
Cell Services:
Cell Line Testing and Assays:

